ABSTRACT
To determine the molecular profile of the multidrug-resistant (MDR) strains of Mycobacterium tuberculosis isolated in Senegal. From 2011 to 2014, 43 MDR strains isolated at the National Reference Laboratory for Mycobacteria of the National Antituberculosis Program (NAP) were characterized by spoligotyping. Nine different spoligotypes were identified, three of which were unique, the other six forming clusters of two to nine isolates. The three principal families identified were the T superfamily (12/43, 27.90%), Beijing (9/43, 20.93%), and Latino-American-Mediterranean (LAM; 3/43, 6.97%). Twelve isolates could not be attributed to a particular lineage. The strains causing multidrug-resistant tuberculosis in Senegal belong to three main families: the T superfamily, Beijing, and LAM.
Key words: Tuberculosis, spoligotypes, resistance, Senegal.
Multidrug resistance to antituberculous drugs is defined as resistance to at least isoniazid (INH) and rifampicin (RMP) (Affolabi et al., 2009). In Senegal, the rate of multidrug resistance to antituberculous drugs in 2016 was 0.9% for new cases and 19% for patients previously treated for the infection, for a total of 13,117 cases of tuberculosis (WHO, 2017). Tuberculosis is thus a major public health problem in Senegal. As the worldwide emergence of multidrug-resistant strains has become one of the greatest public health concerns, understanding the genetic backgrounds of circulating drug-resistant strains is crucial (Ejo et al., 2015). Molecular epidemiology techniques could help us to understand the dynamics of tuberculosis transmission (Affolabi et al., 2009). Several PCR-based methods have been developed to discriminate among strains, including spoligotyping which relies on analyzing the polymorphism of 43 unique DNA sequences comprising identical 36-bp fragments that are repeated in the direct repeat region of the mycobacterial genome (Kamerbeek et al., 1997). Unfortunately, as in many African countries, few molecular typing data are available in Senegal for the strains involved. In this study, the molecular profiles of multidrug-resistant (MDR) mycobacterial strains isolated over a four-year period in Senegal, to bridge this gap was determined.
Patients and strains
Isolates obtained at the National Reference Laboratory for Mycobacteria of the National Antituberculosis Program (NAP) in Senegal was worked on. These strains were isolated between January 2011 and December 2014, mostly from patients experiencing treatment failure or relapse, from Dakar and other regions of Senegal. The clinical data were collated from analysis reports and included the following items: surname, first name, age, sex, clinical diagnosis, history of treatment with antituberculous drugs and the healthcare structure of origin.
Culture and tests of susceptibility to antituberculous drugs (drug susceptibility tests, DSTs)
DSTs were performed at the reference laboratory of the NAP in Senegal. Isolates were cultured on Lowenstein-Jensen (LJ) medium or on LJ medium supplemented with pyruvate. The strains were identified on the basis of the time taken for the colonies to appear, the results of the niacin test, and tests for heat-labile catalase (at 22 and 70°C) and for nitrate reductase. Depending on the availability of reagents, the DSTs were performed by the proportion method (Canetti et al., 1963) or by molecular methods. For the determination of antibiograms by the proportion method, the samples were decontaminated with 4% NaOH and centrifuged. The resulting pellet was then used to inoculate plain LJ medium, LJ medium supplemented with pyruvate and LJ medium supplemented with 5 mg/ml thiophene-2-carboxylic acid hydrazide (TCH). The molecules tested were: ethambutol (EMB, at a concentration of 2 µg/L), streptomycin (SM, 4 µg/L), isoniazid (INH, 2 µg/L) and rifampicin (RMP, 40 µg/L). A strain was considered to be resistant to a drug if its growth in media containing this drug was ≥1% that of the control. Molecular DSTs were based on two tests recommended by the WHO (Find, 2015; WHO, 2011) for identifying mycobacteria of the tuberculosis complex and the mutations most frequently conferring resistance to RMP and INH: the line probe assay (LPA) (Find, 2015) and the Xpert MTB/RIF assay (WHO, 2011). The samples were thus tested with the Xpert MTB/RIF assay, the LPA, or both. In cases in which both tests were used, the LPA was used, above all, to check the susceptibility to INH of strains resistant to RMP in the Xpert MTB/RIF assay.
Molecular typing of strains
This part of the work was carried out in the reference laboratory for mycobacteria in Cotonou, Benin. Spoligotyping was used for molecular characterization. The standardized method was used as described by Kamerbeek et al. (1997), based on the detection of polymorphism of the direct repeat (DR) region. This molecular typing method is based on the polymorphism of nucleotide sequences located between the identical 36-base pair sequences of the DR region present only in members of the M. tuberculosis complex. The number of DR sequences can vary between strains of the same species. These 36 bp sequences are separated by 36-41 bp non-repetitive DNA sequences known as inter-DR sequences or spacers, which display limited variation. Forty three spacer sequences were synthesized and immobilized on a commercial nylon membrane (Isogen Bioscience B.V., BT Maarsen, the Netherlands). Two primers complementary were used to conserve part of the DR region, directed towards the exterior, to amplify the spacers. Genomic profiles were determined by hybridizing the membrane bearing all 43 spacer sequences with these probes. Spoligotypes were then defined on the basis of the presence or absence of several spacers.
Ethical considerations
This retrospective study included data collected during routine diagnosis and treatment, so it did not require ethics committee approval.
Data analysis
The data were entered into the computer and analyzed with EPI-INFO version 7 software (Centers for Disease Control and Prevention, Atlanta, GA, United States).
Forty three MDR isolates from 43 patients aged 9 to 59 years were studied. The mean age of the patients was 34.17 years and the sex ratio (M/F) was 1.66. Most of these patients (82.5%) came from the capital of Senegal, Dakar, and they were essentially consulting for a check-up whilst on antituberculous treatment (37.2%), treatment failure (23.25%) or relapse (16.27%). Sixteen isolates (37.21%) were tested by the proportion method. Two of these isolates were susceptible to EMB and another two were susceptible to SM. The other 27 strains were tested with the Xpert MTB/RIF test, the LPA, or both. Nine different spoligotypes were identified, three of which were unique, whereas the other six formed clusters of two to nine isolates (Table 1). The two principal clusters were ST53 and ST1, which contained twelve and nine isolates, respectively. The three principal families found were the T superfamily (12/43, 27.9%), Beijing (9/43, 20.93%) and LAM (3/43, 6.97%) (Table 1 and Figure 1). Twelve isolates could not be attributed to any particular lineage (Table 1).
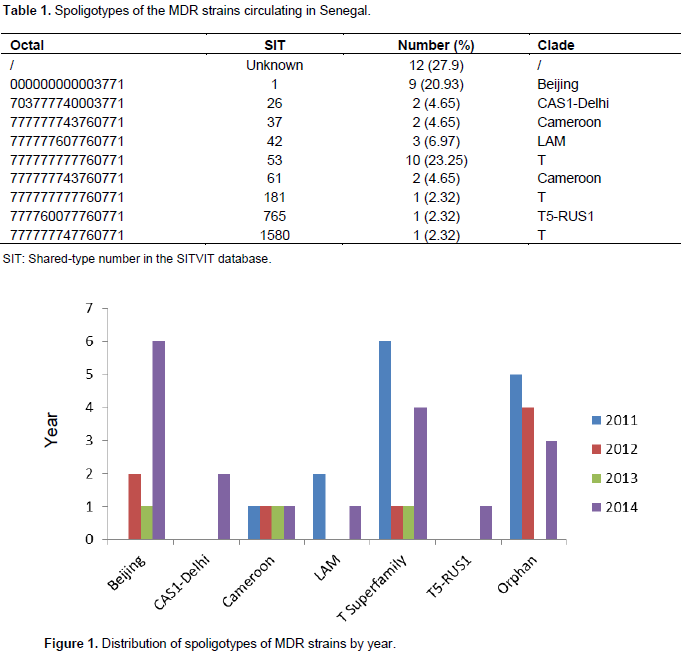
This study is one of the first to investigate the molecular profile of the mycobacterial strains circulating in Senegal and it is the first to focus on the molecular profile of MDR strains in particular. Only two other studies have investigated the spoligotypes circulating in Senegal (Niang et al., 1999; Diallo et al., 2016). Data analysis and comparison with international databases, particularly, the SITVIT2 and SpolDB3 databases identified two major spoligotypes: ST53 and ST1, corresponding to the T superfamily and the Beijing family, respectively. The T superfamily remains poorly defined (Brudey et al., 2006). It is ubiquitous (Brudey et al., 2006), but more frequent in Europe than elsewhere (Bezanahary et al., 2008). In Guadeloupe, if all strains (MDR and non-MDR) are considered together, the T superfamily is also the major family identified (Brudey et al., 2006). By contrast, in the French territories in the Americas (Guadeloupe, Martinique and French Guiana), the X and Latino-American-Mediterranean (LAM) lineages predominate among resistant and MDR isolates, accounting for 10.5 and 42.3% of MDR isolates, respectively (Millet et al., 2014). However, the four principal spoligotypes associated with antituberculous drug resistance in French territories in the Americas (SITs 20, 15, 46, and 64) (Millet et al., 2014) are different from those identified in the present study.
In Burkina Faso, most isolates belong to the LAM (30%), T (20%) and Haarlem (9%) families (Godreuil et al., 2007). The second major family of MDR strains identified in the present study was the Beijing family. The strains of this family, the most widespread family worldwide (Arora et al., 2014), are known to be associated with multidrug resistance to antituberculous drugs (European Concerted Action, 2006; Narvskaia et al., 2002). Furthermore, their rapid propagation, for multiple reasons (demographic changes and globalization), in certain contexts, suggests that the members of this family are intrinsically virulent (Affolabi et al., 2009; Bezanahary et al., 2008). The presence of strains from this family in Senegal may be linked, as previously suggested (Bezanahary et al., 2008), to immigrants originating from the Far East. The percentage of Beijing strains identified in this study (20.45%) is greater than that found in Benin (10.3%). All the Beijing isolates in the present study were obtained in the region of Dakar, the capital city, which is located on the coast. Otherwise, strains of the Beijing family have rarely been isolated in the coastal countries of West Africa (Godreuil et al., 2007; Cadmus et al., 2006; Niobe Eyangoh et al., 2003). By contrast, in Kyrgyzstan, 13 of the 15 MDR strains detected in prison populations belonged to the Beijing family (Mokrousov et al., 2009).
The third most frequent spoligotype among our multiresistant isolates was ST42, which belongs to the Latin American and Mediterranean (LAM) family. Like ST53 (T superfamily), ST42 is ubiquitous, but more frequent in Europe than elsewhere (Brudey et al., 2006; Bezanahary et al., 2008). The LAM family originated in Latin America and the Mediterranean Basin (Brudey et al., 2003). Its presence in Africa reflects the impact of colonization and past migration (Brudey et al., 2006). The strains of the LAM family are often associated with resistance. Indeed, in the French territories in the Americas (Guadeloupe, Martinique and French Guiana), the LAM lineage is overrepresented among MDR strains with respect to strains with other resistance profiles and susceptible strains (Millet et al., 2014). ST61 has been described as prevalent in the coastal countries of West Africa (Affolabi et al., 2009), but accounted for only 4.55% of the MDR isolates. It does not therefore seem to be strongly linked to multidrug resistance to antituberculous drugs in Senegal. This spoligotype also accounts for the majority of MDR strains in Burkina Faso and Cameroon, where it has been referred to as the “Cameroon family” (Godreuil et al., 2007). Two spoligotypes rarely found in West Africa were detected: ST765 (2.27%) and ST26 (4.55%) corresponding to the T5-RUS1 and Central Asia (CAS1-Delhi) types. These two families were detected in this study in 2014, but were absent between 2011 and 2013.
Type T5-RUS1 (ST765), previously known as “non-LAM families (T1 or T5-RUS)” and recently reclassified as belonging to the LAM family (Abadia et al., 2010; Gibson et al., 2008) originated in the European part of Russia (Mokrousov et al., 2014). The Central Asia type (CAS1-Delhi) may be geographically linked to North India or Pakistan, or to other countries or regions, such as Sudan, Libya or East Africa (Bezanahary et al., 2008). In a pediatric population in India, the CAS1-Delhi type was second in frequency only to the Beijing type among susceptible and MDR isolates (Arora et al., 2014). A large number of MDR isolates in this study were grouped into clusters (28/43 or 65.11%). In total, six clusters were identified, the two principal clusters being ST53 (n=10) and ST1 (n=9). The clustering of isolates in a given population is thought to reflect recent transmission. The other cases, corresponding to strains with a distinctive genomic fingerprint, are thought to result from the reactivation of older infections (Alland et al., 1994; Small et al., 1994). However, the association of spoligotyping with a more discriminatory method, such as MIRU-VNTR, is recommended to improve the study of clusters (Affolabi et al., 2009).
This study provided a snapshot of the spoligotypes of M. tuberculosis circulating in Senegal, including spoligotypes of MDR strains. Three families accounted for most of the MDR strains in Senegal: the T superfamily, Beijing, and LAM. Spoligotypes generally considered to be “European” or “Asian” were found among the MDR isolates: T5-RUS1 and CAS1-Delhi. A large study including susceptible strains and covering the entire country would provide a clearer idea of the molecular profiles of the M. tuberculosis strains circulating in Senegal. This study should combine the use of two techniques: spoligotyping and MIRU-VNTR (Sola et al., 2003).
The authors have not declared any conflict of interests.
The authors thank all the staff of the Reference Laboratory for Mycobacteria in Cotonou (Benin) and the staff of the National Antituberculosis Program (NAP) in Senegal, including, in particular, the staff of the NAP laboratory.
REFERENCES
|
Abadia E, Zhang J, dos Vultos T, Ritacco V, Kremer K (2010). Resolving lineage assignation on Mycobacterium tuberculosis clinical isolates classified by spoligotyping with a new high-throughput 3R SNPs based method. Infect. Genet. Evol. 10:1066-1074.
Crossref
|
|
|
|
Affolabi D, Anyo G, Faïhun F, Sanoussi N, Shamputa IC, Rigouts L, et al (2009). First molecular epidemiological study of tuberculosis in Benin. Int. J. Tuberc. Lung. Dis. 13: 317-322.
|
|
|
|
|
Alland D, Kalkut GE, Moss AR, McAdam RA, Hahn JA, Bosworth W (1994). Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods, N. Engl. J. Med. 16: 1710-1716.
Crossref
|
|
|
|
|
Arora J, Sidiq Z, Sharma S, Singhal R, Bhalla M, Couvin D, Sarin R, Rastogi N, Myneedu VP (2014). Phylogenetic associations with drug-resistant Mycobacterium tuberculosis isolates in a paediatric population, Int. J. Tuberc Lung Dis. 18:1172-1179.
Crossref
|
|
|
|
|
Bezanahary H, Baclet MC, Sola C, Gazaille V, Turlure P, Weinbreck P, Denis F, Martin C (2008). Molecular strain typing contribution to epidemiology of tuberculosis in Limousin (1998 to 2006). Med. Mal. Infect. 38:309-317.
Crossref
|
|
|
|
|
Brudey K, Filliol I, Sola C, Bebear C, Elia-Pasquet S, Texier- Maugein J, Rastogi N (2003). Molecular characterization and biodiversity of Mycobacterium tuberculosis in French overseas departments (French West-Indies and French Guiana) as compared to an area from metropolitan France (Aquitaine). Pathol. Biol. 51:282-9.
Crossref
|
|
|
|
|
Brudey K, Filliol I, Théodore M, Sola C, Rastogi N (2006). Molecular epidemiology of tuberculosis in Guadeloupe from 1994 to 2000. Pathol. Biol. 54:14-21.
Crossref
|
|
|
|
|
Cadmus S, Palmer S, Okker M (2006). Molecular analysis of human and bovine tubercle bacilli from a local setting in Nigeria. J. Clin. Microbiol. 44: 29-34.
Crossref
|
|
|
|
|
Canetti G, Rist N, Grosset J (1963). Mesure de la sensibilité du bacille tuberculeux aux drogues antibacillaires par la méthode des proportions, Rev. Tuberc. Pneumol. 27:217-272.
|
|
|
|
|
Diallo AB, Walbang OG, Camara M, Lo S, Ayorinde A, Niang A, et al (2016). Molecular genotypes of Mycobacterium tuberculosis strains circulating in Dakar, Senegal. Afr. J. Microbiol. Res. 10(35):1460-1466.
Crossref
|
|
|
|
|
Ejo M, Gehre F, Barry MD, Sow O, Bah NM, Camara M et al (2015). First insights into circulating Mycobacterium tuberculosis complex lineages and drug resistance in Guinea. Infect. Genet. Evol. 33:314 - 319.
Crossref
|
|
|
|
|
European Concerted Action on New Generation Genetic Markers and Techniques for the Epidemiology and Control of Tuberculosis (2006). Beijing/W genotype Mycobacterium tuberculosis and drug resistance, Emerg. Infect. Dis. 12(5):736-743.
Crossref
|
|
|
|
|
FIND (2015). Line probe assay (1st line drugs).
|
|
|
|
|
Gibson AL, Huard RC, Gey van Pittius NC, Lazzarini LC, Driscoll J (2008). Application of sensitive and specific molecular methods to uncover global dissemination of the major RDRio Sublineage of the Latin American-Mediterranean Mycobacterium tuberculosis spoligotype family. J. Clin. Microbiol. 46:1259-1267.
Crossref
|
|
|
|
|
Godreuil S, Torrea G, Terru D, Chevenet F, Diagbouga S, Supply P, Van de Perre P, Carriere C, Banuls AL (2007). First Molecular Epidemiology Study of Mycobacterium tuberculosis in Burkina Faso, J. Clin. Microbiol. 45:921-927.
Crossref
|
|
|
|
|
Kamerbeek J, Schouls L, Kolk A, Van Agterveld M, Van Soolingen D, Kuijper S (1997). Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914.
|
|
|
|
|
Millet J, Berchel M, Prudenté F, Streit E, Bomer AG, Schuster F, Vanhomwegen J, Paasch D, Galbert I, Valery E, Aga R, Rastogi N (2014). Resistance to First-Line Drugs and Major Genotypic Lineages of Mycobacterium tuberculosis in the 3 French Department of the Americas: Profiles, Evolution, and Trends (1995–2011). Bull. Soc. Pathol. Exot. pp. 2-16.
|
|
|
|
|
Mokrousov I, Valcheva V, Sovhozova N, Aldashev A, Rastogi N (2009). Penitentiary population of Mycobacterium tuberculosis in Kyrgyzstan: exceptionally high prevalence of the Beijing genotype and its Russia-specific subtype. Infect. Genet. Evol. 9:1400-1405.
Crossref
|
|
|
|
|
Mokrousov I, Vyazovaya A, Narvskaya O (2014). Mycobacterium tuberculosis Latin American-Mediterranean family and its sublineages in the light of robust evolutionary markers. J. Bacteriol. 196:1833-41.
Crossref
|
|
|
|
|
Narvskaia OV, Mokrousov IV, Limeshchenko EV, Steklova lN, Otten TF, VishnevskiÄ BI (2002). Characterization of Mycobacterium tuberculosis strains prevalent in North-West of Russia by spoligotyping, Probl. Tuberk. 4:44-48.
|
|
|
|
|
Niang MN, De la Salmoniere YG, Samb A, Hane AA, Cisse MF, Gicquel B, Perraut R (1999). Characterization of M. tuberculosis strains from west African patients by spoligotyping, Microbes. Infect. 1:1189-1192.
Crossref
|
|
|
|
|
Niobe-Eyangoh SN, Kuaban C, Sorlin P (2003). Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J. Clin. Microbiol. 41:2547-2553.
Crossref
|
|
|
|
|
Small PM, Hopewell PC, Singh SP, Paz A, Parsonnet J, Ruston DC (1994). The epidemiology of tuberculosis in San Francisco. A population based study using conventional and molecular methods, N. Engl. J. Med. 16: 1703-1709.
Crossref
|
|
|
|
|
Sola C, Filliol I, Legrand E, Lesjean S, Locht C, Supply P, Rastogi N (2003). Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics, Infect. Genet. Evol. 3:125-133.
Crossref
|
|
|
|
|
World Health Organization WHO (2011). Rapid implementation of Xpert MTB/RIF. WHO Report, Geneva. WHO/HTM/TB/ 2011.2, 2011.
|
|
|
|
|
World Health Organization (WHO) (2017). Tuberculosis Country profiles: Senegal.
View Accessed February 2018.
|
|