Short Communication
ABSTRACT
A variant is an alternative nucleotide located at a specific region of a gene. 48 genes encode for human red cell blood group systems. Variants within these genes encode for alleles, which can be highly polymorphic. The blood group gene loci jointly display all types of inherited variants to include single nucleotide variants (SNVs), insertions/deletions (indels), and structural variants. In Africa, there is limited information on the red cell variants. The aim of the study was to determine the incidence of Kell red blood cell variants among the Kenyans. Blood donor’s samples that were used for routine grouping process were identified for this study. Next generation sequencing method was employed to predict the Kell red cell variants in blood donor samples. Descriptive statistics was applied and the result was presented in form of tables. The study reveals that Kell system has six variants with two major phenotypes and genotpes. The most common phenotype genotyped was KEL2 (KEL*02/KEL*02) 79% (85/108) followed by KEL2 KEL6 KEL7 (KEL*02.06/KEL*02) 16.7% (18/108). The rest were found to be of low frequency and all were associated with KEL2. The study recommends an extended study with a large sample size and introduction of extended phenotyping to aid Kell antigens identification in the donor population.
Key words: Kell, variant, allele, KEL, KEL*02/KEL*02, molecular, genome, gene, next generation sequencing.
Abbreviation: EDTA, Ethylenediaminetetraacetic acid; HDFN, haemolytic disease of the fetus and the new born; HTR, haemolytic transfusion reaction; KNBTS, Kenya National Blood Transfusion Service; RBCs, red blood cells; SNVs, single nucleotide variants; MKU, Mount Kenya University; NGS, next generation sequencing.INTRODUCTION
A variant is an alternative nucleotide whose location is at a specific region within a gene or genome. A gene variant is a permanent change in the DNA sequence that makes up a gene. 48 genes encode for human red cell blood group systems. Variants within these genes encode for alleles, which can be highly polymorphic. The blood group gene loci jointly display all types of inherited variants to include single nucleotide variants (SNVs), insertions/ deletions (indels), and structural variants (Johnsen, 2015).
The aim of the study was to determine the incidence of Kell red blood cell variants among the blood donor’s population of Kenya.
Kell blood group system is a diverse glycoprotein membrane located on chromosome 7q33 and its manifestation arises in hematopoietic (erythroid) cells. It is given ISBT symbol: KEL, number 006 and gene symbol KEL (Dean, 2005). It is under the control of KEL gene encrypted by 19 exons the polymorphism of Kell is associated with a point alteration (mutations) in a specific region of the genome. It also exhibits a rare null (Ko) type that lacks all expression of Kell antigens on the red cells. The Kell variants and aggulutinogens are of clinical importance in the transfusion medicine. They have been defined as strong triggers of immunological reactions when introduced to those who lack the specific agglutinations. They have been implicated in severe HDFN and acute red cell destructions in transfusions therapy.
In the white population, Kell is categorized as one of the three clinical significant blood system after ABO and Rh blood groups (Liu et al., 2012).
The Kell systems encompass over 34 aggulutinogens. The major types are K and k (Cellano), KEL*01 and KEL*02.The K antigen (KEL*01) is of clinical significance among the two aggulutinogens. Anti-K is associated with serious newborn anemia because it inhibits production of red blood cells by the stem cells in the bone marrow (erythropoiesis) newborn resulting to low count of platelets and neutrophils (thrombocytopenia and neutropenia) (Guelsin et al., 2011).
MATERIALS AND METHODS
A total of 108 anonymous left over blood samples collected in EDTA vacutainers were identified, selected and prepared for molecular genotyping. Manual Genomic DNA extraction was accomplished by using QIAamp whole blood DNA mini kit (250 51106) as per manufacturer’s instructions (Qaigen Germany). Library preparation was performed using a custom blood grouping enrichment panel (Roulis et al., 2020), and the Illumina DNA prep Enrichment protocol as per manufacturer’s instructions (Illumina Inc., San Diego, CA, USA).
Inclusion criteria
Samples collected in 4 ml ETDA tubes that were non-reactive for transfusion transmission infections (TTIs), with no haemolysis.
Exclusion criteria
All samples that failed the required inclusion criteria in preparation for the manual DNA extraction.
RESULTS
The study reveals that Kell system has six variants with two major phenotypes and genotpes.The most common phenotype was KEL2 (KEL*02/KEL*02)79%(85/108) followed by KEL2 KEL6 KEL7 (KEL*02.06/KEL*02) 16.7%(18/108).The rest were found to be of low frequency and all were associated with KEL2 as shown Table 1.
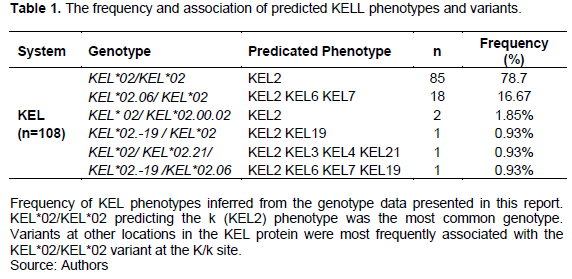
DISCUSSION
The study reveals that Kell system has six variants and their predicted phenotypes. Most common was KEL2 (KEL*02/KEL*02) 85 (79%) followed by KEL2 KEL6 KEL7 (KEL*02.06/KEL*02) 18 (16.7%).The rest were found to be of low frequency 2-1% and all were associated with KEL2 (k+).
The study outcome has shown that indeed there is heterogeneity in the distribution patterns of Kell variants and phenotypes globally. It has also shown that k+ is the most occurring in the donor population of Kenya. This k+ is implicated in red cell alloimmunisation, thrombocytopenia, and fetomaternal hemorrhage, new natal and infants breeding and organ rejections, thus it is of clinical significance in the transfusion and transplantation therapy. Having this information is very important for supporting resolving complex cases associated with red cell therapy. Comparing the results with similar studies, there is variance in different populations as observed by Dean (2005). KEL2 (k+) was found to be 91% in the white population, and 98% in the people of black colour. KEL1 (K) was recorded as having a low frequency of 0.2% in whites and very rare in the dark population. KEL1/KEL2 (K+k+) 8.8% in whites and 2% in people of black colour (Dean, 2005).
In the Chinese community, K (KEL1) frequency was 3.5% and k (KEL2) 99.97% (Liu et al., 2012). In Indian study, KEL2 (k+) frequency is 96.5%, whites is 91% and dark colour (blacks) is 98% and 100% in Chinese populace (Makroo et al., 2013). In Northern India, the incidence of Kell types have been shown as KEL1 (K) 2.57% whereas KEL2 (k+) as 97.43% (Mangwana et al., 2021; Thakral et al., 2010).
In the Japanese population, the study revealed KEL*01/KEL*02 as 0.48%; KEL*02/KEL*02 as 99.52%, KEL*02 as 99.76% and KEL*01.01 as 0.024% (Flôres et al., 2013). Study from Saudi Arabia indicates that KEL2 (k+ Cellano) was the most common (100%) whereas KEL1 (K) recorded 8% (Amani et al., 2020).
A study from Morocco, KEL1 (K) is 0.19% whereas KEL2 is 99.80% (Benahadi et al., 2014). Similar study conducted in Southern Brazil documented the frequencies as KEL*02/KEL*02: 94 (75%), KEL*01/KEL*02: 5.0%, while KEL*01/KEL*01 was 0.25% (Guelsin et al., 2011; Flôres et al., 2014).
CONCLUSION
The study has revealed the organization of Kell red cell variants in the donor population of Kenya. It has also shown that k+ (KEL2) is the most prevalent phenotype. These findings are also the first of this kind in Kenya. In addition to confirmed presence of Kell red cell variants, valuable knowledge is presented on the basis for improved investigations of these variants. The generated evidence is useful in forming a base for the preparation of red cell antigen panels, advocacy for the initiation for extended phenotyping and in the generation of a red cell genomic library/gene reference for Kenya and her neighboring countries.
RECOMMENDATION
Considering the clinical implications associated with Kell antigens (transfusion reactions, infants breeding, thrombocytopenia and organ rejections); this outcome is very beneficial in resolving cases associated with adverse transfusion reaction especially in ongoing and massively transfused patients (Sickle cells, cancer cases among others) matching and as a base for a reference gene bank in Kenya. The results of this study in combination with others related to other blood groups red cell variants will be useful in instituting a blood grouping molecular reference laboratory in Kenya and support for East Africa community. It will also be useful evidence while considering the use of extended phenotyping in blood bank and tissue transplant laboratories both in Kenya and East Africa in the identification of rare phenotypes that are of clinical significance in transfusion and transplantation medicine. More research on this field in Kenya and East Africa will augment the gene data base and support the blood grouping laboratories in the investigations of complicated complex cases as well as enhance collaborations nationally and internationally.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
ACKNOWLEDGEMENTS
The authors thank the Ministry of Health-Kenya, Tissue and Transplant Authority Kenya (formally Kenya National Blood Transfusion Service), Mount Kenya University, Queensland University of Technology Australia, and Australian Red Cross Blood Life.
ABBREVIATIONS
EDTA, Ethylenediaminetetraacetic acid; HDFN, haemolytic disease of the fetus and the new born; HTR, haemolytic transfusion reaction; KNBTS, Kenya National Blood Transfusion Service; RBCs, red blood cells; SNVs, single nucleotide variants; MKU, Mount Kenya University; NGS, next generation sequencing.
REFERENCES
|
Amani YN, Alumran NA, Alzahrani F (2020). Phenotype frequencies of major blood group systems (Rh, Kell, Kidd, Duffy, MNS, P, Lewis, and Lutheran) among blood donors in the Eastern Region of Saudi Arabia. Journal of Blood Medicine 11:59-65. |
|
|
|
|
|
Benahadi A, Boulahdid S, Adouani B, Laouina A, Mokhtari A, Soulaymani A, Hajjout K, Benajiba M, Alami R (2014). Mapping rare erythrocyte phenotypes in Morocco: A tool to overcome transfusion challenges. Journal of Blood Transfusion 2014:e707152 |
|
|
|
|
|
Dean L (2005). The Duffy blood group. In Blood Groups and Red Cell Antigens [Internet].National Center for Biotechnology Information (US). Available at: |
|
|
|
|
|
Flôres MA, Vientiane JEL, Guelsin GAS, Fracasso AS, Melo FC, Hashimoto MN, Sell AM (2014). Rh, Kell, Duffy, Kidd and Diego blood group system polymorphism in Brazilian Japanese descendants. Transfusion and Apheresis, Science 50(1):123-128. |
|
|
|
|
|
Guelsin GAS, Sell AM, Castilho L, Masaki VL, Melo FC, Hashimoto MN, Hirle LS, Visentainer JEL (2011). Genetic polymorphisms of Rh, Kell, Duffy and Kidd systems in a population from the State of Paraná, southern Brazil. Revista Brasileira de Hematologia e Hemoterapia 33(1):21-25. |
|
|
|
|
|
Johnsen JM (2015). Using red blood cell genomics in transfusion medicine. Hematology 2015(1):168-176. |
|
|
|
|
|
Liu Z, Zeng R, Chen Q, Li M., Shi G, Wei P, Huang C, Tang R, Sun J, Zhang X (2012). Genotyping for Kidd, Kell, Duffy, Scianna, and RHCE blood group antigens polymorphisms in Jiangsu Chinese Han. Chinese Medical Journal 125(6):1076. Available at: |
|
|
|
|
|
Makroo RN, Bhatia A, Gupta R., Phillip J (2013). Prevalence of Rh, Duffy, Kell, Kidd and MNSs blood group antigens in the Indian blood donor population. The Indian Journal of Medical Research 137(3):521-526. Available at: |
|
|
|
|
|
Mangwana S, Simon N, Sangwan L (2021). RH phenotype, ABO and Kell antigens, alleles and haplotypes frequencies in North Indian blood donor population. Global Journal of Transfusion Medicine 6(1):81. Available at: |
|
|
|
|
|
Roulis E, Schoeman E, Hobbs M, Jones G, Burton M, Pahn G, Liew YW, Flower R, Hyland C (2020). Targeted exome sequencing designed for blood group, platelet, and neutrophil antigen investigations: Proof-of-principle study for a customized single-test system. Transfusion 60(9):2108-2120. |
|
|
|
|
|
Thakral B, Saluja K, Sharma RR, Marwaha N (2010). Phenotype frequencies of blood group systems (Rh, Kell, Kidd, Duffy, MNS, P, Lewis, and Lutheran) in north Indian blood donors. Transfusion and Apheresis Science 43(1):17-22. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0