ABSTRACT
The most conserved regions for 15 hepatitis B virus complete genome of different subtypes were aligned using PCGENE software CLUSTAL to design a new pair of primer that can bind to each subtype of hepatitis B virus (HBV), to amplify PreS region of HBV genome. A pair of primer from these conserved patches was selected using software PRIMER and named as Nhepf1 and Nhepr1. Nhepf1, forward primer bound 2362-2385 nucleotides and Nhepr1, reverse primer bound 260-283 nucleotide amplify 1.12 Kb region of HBV genome that contain PreS sequence. The pair of primer was optimized for PCR. Nhepf1 and Nhepr1 annealed well at 50°C to subtype adw2 (American), adr4 (Japanese) and Pakistanian patient derived HBV DNA without any nonspecific bands. The results were found to be highly reproducible with greater accuracy.
Key words: Hepatitis B virus (HBV) genome, HBV conserved region, polymerase chains reaction (PCR) primers, primer designing.
There is prevalence of hepatitis B virus in an estimated 500 million person in the world, the frequent occurrence of this infection in close contacts of carriers, perinatal vertical transmission from mother to infant and high incidence of insidious chronic liver disease or cancer of the liver among the carriers, signify a major public health problem of worldwide concern (Abbas, 2007). Hepatitis B viral genome (3200 bp) has four known genes, S encoding the viral surface protein, C encoding core protein, P DNA polymerase encoding gene and X gene (Lai et al., 1991). Virus envelope proteins designated PreS1, PreS2 and S carries neutralization epitopes and plays an important role in virus/cell interactions (Wong et al., 2004). At least five epitopes have been mapped within PreS region (Kuroki et al., 1990) and it contains two directly or indirectly binding sites to hepatocytes that constitute the first step in the infection (Dash et al., 1992). Antibodies to PreS region are considered to play a major role in clearance of HBV. Various naturally occurring mutations in the PreS and S genes have been described, including deletions and point mutations leading to subtypic changes (Okamoto et al., 1987 and Lai et al., 1991).
There are at least four subtypes (adw, adr, ayr and ayw) of HBV that can be recognized by antigenic differences in the antigen (Ono et al., 1983; Abbas et al., 2006; Abbas, 2009). HBsAg has distinct geographical, epidemiological and anthropological settings (Courouce-Pauty et al., 1983; Abbas et al., 2008). The considerable number of HBV isolates with divergent nucleotide sequences and the partially double stranded charac-teristic of HBV impose the need for extreme care in the choice of primers for both full length and fragment amplification (Zhang et al., 2007).
Subtypes provide useful epidemiological marker of HBV. The mutation rate of HBV is estimated to be 100 times higher than that of other DNA viruses. Since HBV infection persists for many decades in patients, base changes can accumulate over time (Drosten et al., 2000). One in every ten HBV isolates is slightly different at the nucleotide level from other all sequenced isolates, that is, >10% sequence heterogeneity in HBV (Lauder et al., 1993; Zhang et al., 2007). It is hence crucial to study sequence variation of envelope protein of Pakistanian HBV isolates and to plan a noval strategy for designing a new set of primers with the anticipated use for clinical diagnosis of HBV after the selection of the most conserved region of HBV genomic DNA of different subtypes. The aim of the current investigation was to design such a pair of primer which could perfectly bind to all different genotypes and subtypes of HBV, and would be helpful in detection and DNA amplification of HBV isolated from areas (as Pakistan) that have no previous HBV sequence data.
Primer designing
Eight sequences were collected from GenBank and seven were selected and edited from research papers and fed into computer (PCGENE Release 6.5, 1991 Intelligenetics Inc.). Two sets of subfiles, for each of the fifteen sequences, one having 200 nucleotides stretch from 2300/2307 to 2500/2507 positions and other of 300 nucleotides stretch from 155/157 to 455/457 positions, were created using SEQIN (PCGENE). Fifteen subfiles for each portion were compiled in a library file using FILE (PCGENE). These library files were used in CLUSTAL (PCGENE), to align fifteen patches of HBV genome. The most conserved regions found in this way were subjected to biological software PRIMER version 0.5 (MIT USA, 1991) for primer selection.
DNA preparation
An American clone adw2 gifted by Dr. Aleem Siddique (NIH USA) and a Japanese clone kindly provided by Dr. Okubo (Japan) were prepared as in Sambrook et al. (1989). HBV DNA from sera of HbsAg positive patients were extracted as described in Abbas et al. (2005).
PCR amplification
The cloned DNA as well as patient derived HBV DNA were used for PCR amplification using 1 mM of each Nhepf1 and Nhepr1 with 10 mM Tris-Cl (pH 8.3), 2.5 mM MgCl2, 50 mM KCl, 200 mM each dNTP, 2U Taq polymerase per 100 ml reaction (Persing et al., 1993). The mixture was denatured at 95°C for 2 min, annealed at 50°C for 2 min, and elongated at 70°C for 2 min for 35 cycles. PCR product was resolved on 1% agarose gel according to Sambrook et al. (1989).
Fifteen (15) sequences of different genotypes and subtypes were collected and aligned to find conserved regions to design a pair of primer. Eight nucleotide sequences of hepatitis B virus were collected from Genbank with accession numbers were M32138, X75658, X75657, X04615, X75664, X75665, X75663, X75656 and seven selected from research papers (Okamoto et al., 1986; Norder et al., 1994; Tong et al., 1990; Ono et al., 1983). Two regions which were reported to be conserved (Lauder et al., 1993) were selected. The 200 nucleotides conserved patch was from the end of core region overlapping with start of P gene at 2300/2307-2500/2507 position. This patch gave only one region of 37 nucleotides (Figure 1) after alignment of library file through CLUSTAL, which was adopted for forward primer selection.
The 300 nucleotide conserved patch selected for reverse primer was from first hydrophobic region of surface antigen at 155/157-455/457 position. The alignment of library file through CLUSTAL of this patch gave three stretches of 27 nucleotides each (Figure 2). First stretch was chosen for reverse primer selection.
The above two selected regions which were found absolutely conserved and consensus to each other by CLUSTAL were used to select forward and reverse primer respectively by PRIMER selection (Figure 3). After examining 37 forward primers it accepted 9 forward primers, similarly after examining 27 reverse primers, it accepted 10 reverse primers. Finally, one pair of 24-mer each was selected AS given below:
Nhepf1 GTCCCCTAGAAGAAGAACTCCCTC Tm= 62.0°C
Nhepr1 CCCTAGAAAATTGAGAGAAGTCCA Tm= 60.1°C
Forward primer possesses one GC clamp while reverse primer possesses two GC clamps at its 3’ end. GC contents of primers were 54%. Both primers have five self-complementary bases (2.83%), which were not consecutive to each other but


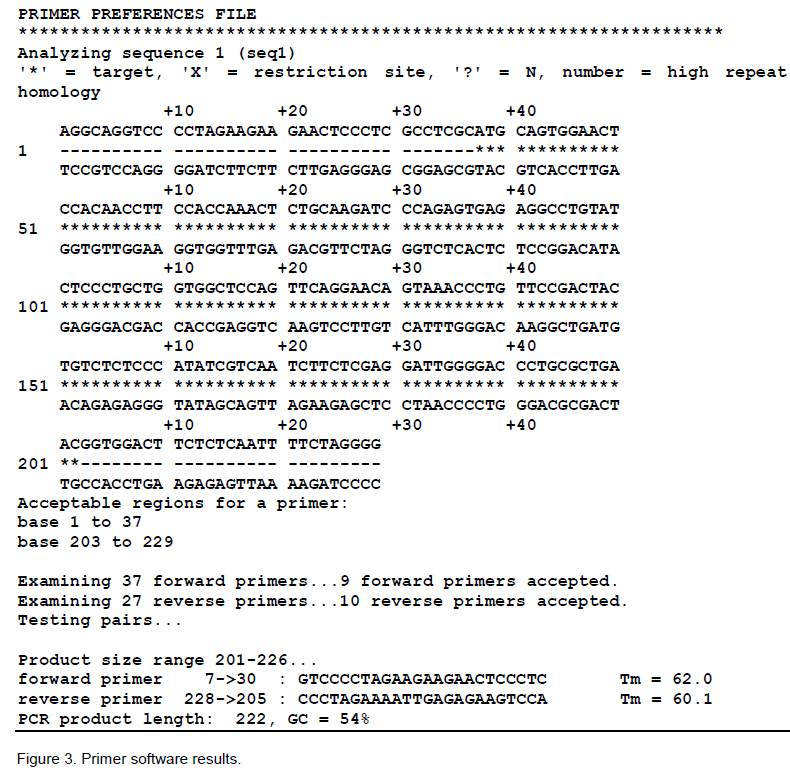
occur at irregular intervals. Tm were 62 and 60.8°C, respectively. The annealing temperature of the primers is 10-15° below the Tm of the primers (Desselberger, 1995). Therefore, PCR was tried using newly selected primers at different annealing temperatures ranging from 60-48°C. The optimum temperature selected was 50°C, which was then used for routine PCR. DNA concentrations for cloned adw2 and adr4 were tried from 10 - 500 ng, 50 ng was found best. MgCl2 concentration found best for these primers was 1.5 mM of tested ranges 1-4 mM. Primer (NhepF1 and NhepR1) amplified a fragment of 1.1 Kb of PreS region both in pSHBV and patient derived HBV DNA (Figure 4). HBV clone pSHBV gave 3.7 Kb as the orientation of the HBV insert, vector was amplified along with insert lane 2 and 4 (Figure 4).
Hepatitis B virus is the smallest known DNA virus that cause infection in man. The genome is partially double

stranded circular, 3.2 Kb DNA molecule, 0.78 mM in length which corresponds to a molecular weight around 1.6 x 106. HBV genome contains an EcoRI site which lies 9 bp downstream from 5’end of PreS2 region. A new set of well conserved primers of 24-mer each were selected from consensus regions (the region encoding hydro-phobic region of HBV surface gene and the start of the P gene which overlaps the end of gene C).
It is also imperative that the primers should be greater than 18 or 20 mer, in the range of 20-25 mer, so that they may provide better background. The primers should have at least two to three GC clamps at their 3’end to have a strong grip during the positioning and walking of Taq DNA polymerase to extend the primer for polymerization. In the ideal condition, the F and R primers should not have any intra or/and inter complementary bases, but if there is, they should not be greater than 2.5%. The complementary bases should not be consecutive to each other but present at irregular intervals, because all the complementary bases at one site are consecutive to each other, more likely have a chance to come close together quickly and complement each other and may have strong grip as compared to the self-complementation at distance intervals.
We look forward with the hope that this pair can be used for routine clinical diagnosis of hepatitis B virus infection in the diversified geographical world of highly variable strains of HBV. This may prove as the reliable primers for obtaining accurate and confident results.
The author(s) have not declared any conflict of interests.
REFERENCES
|
Abbas N, Abbas S, Afroze T (2008). PCR amplification of Hepatitis B Virus in limiting DNA dilution format. Pak. J. Zool. 40(4):255-260.
|
|
|
|
Abbas N, Shakoori AR (2007). Hepatitis B virus X gene. A Review: Proceedings Pak Congress Zool. 27:127-136.
|
|
|
|
|
Abbas N, Rasool M, Afroze T (2005). PCR based diagnosis of hepatitis B virus. Pak. J. Zool. 37(4):285-288.
|
|
|
|
|
Abbas N, Yousra A, Shakoori AR (2006). Mutations in the Hepatitis B virus core gene and its efficacy as a vaccine, A Review: Proceedings Pak Congress Zool. 26:301-319.
|
|
|
|
|
Courouce-Pauty AM, Plancon A, Soulier JP (1983). Distribution of HBsAg subtypes in the world. Vox Sang 44:197-211.
Crossref
|
|
|
|
|
Dash S, Rao KV, Panda SK (1992). Receptor for pre-S1(21-47) component of hepatitis B virus on the liver cell: role in virus cell interaction. J. Med. Virol. 37(2):116-121.
Crossref
|
|
|
|
|
Desselberger U (1995). Medical Virology: A Practical approach. IRL press Oxford University Press New York.
|
|
|
|
|
Drosten C, Weber M, Seifried E, Roth WK (2000). Evaluation of a new PCR assay with competitive internal control sequence for blood donor screening. Transfusion 40(6):718-724.
Crossref
|
|
|
|
|
Kuroki K, Froreani M, Mimms LT, Ganem D (1990). Epitope Mapping of the Pres1 domain of the Hepatitis B virus large surface protein. Virol. 176:620-624.
Crossref
|
|
|
|
|
Lai KE, Melis A, Mazzoleni AP, Farci P, Balestrier A (1991). Sequence analysis of hepatitis B virus genome of new mutant of ayw subtype isolated in Sardinia. NAR, 19:5078.
Crossref
|
|
|
|
|
Lauder IJ, Lin HJ, Lau JYN, Siu TS, Lai CL (1993). The variability of the hepatitis B virus genome: Statistical analysis and biological implications. Mol. Biol. Evol., 10(2):457-470.
|
|
|
|
|
Naaz A (2009). Variability anlysis in Immunogenic Region of Hepatitis B virus: Sequence variation in Pakistanian Hepatitis B virus Variability anlysis in Immunogenic Region of Hepatitis B virus: Sequence variation in Pakistanian Hepatitis B virus. VDM VerlagDr Muller Aktiengesellschaft& Co. KG DudweilerLandstr. Saarbrucken, Germany. 99:66123. ISBN: 978-3-639-14908-1.
|
|
|
|
|
Norder H, Courouce AM, Magnius LO (1994). Complete genomes, phylogenetic relatedness, and strurctural proteins of six strains of the HBV, four of which represent two new genotypes. Virol. 198(2):489-503.
Crossref
|
|
|
|
|
Okamoto H, Imai M, Shimozaki M, Hoshi Y, Iizuka H, Gotanda T, Tsuda F, Miyakawa Y, Mayumi M (1986). Nucleotide sequence of a cloned HBV genome, subtype "ayr": Comparison with genomes of the other three subtypes. J. Gen. Virol. 67(11):2305-2314.
Crossref
|
|
|
|
|
Okamoto H, Imai M, Tsuda F, Tanaka T, Miyakawa Y, Mayumi M (1987). Point mutations in the S gene of HBVfor a d/y or w/r subtypic change in two blood donorscarring a surface antigen of compound subtype adyr or adwr. J. Virol. 61:3030-3034.
|
|
|
|
|
Ono Y, Onda H, Sasada R, Igarashi K, Sugino Y, Nishioka K (1983). The complete nucleotide sequence of the cloned hepatitis B virus DNA: subtype adr and adw. NAR, 11(6):1747-1757.
Crossref
|
|
|
|
|
Persing DH, Smith TF, Tenover FC, White JT (1993). Diagnostic Molecular Biology: Principles and Applications. American Society for Microbiology, Washington D.C.
|
|
|
|
|
Sambrook J, Fritsch EF, Maniatis T (1989). Molecular Cloning: a Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
|
|
|
|
|
Tong SP, LI J, Vitvitski L, Trepo C (1990). Active hepatitis B virus replication in the presence of anti-HBE is associated with viral variants containing an inactive pre-C region. Virol. 176:596-603.
Crossref
|
|
|
|
|
Wong DK, Yuen MF, Tse E, Yuan H, Sum SS, Hui CK, Lai CL (2004). Detection of intrahepatic hepatitis B virus DNA and correlation with hepatic necroinflammation and fibrosis. J. Clin. Microbiol. 42(9):3920-3924.
Crossref
|
|
|
|
|
Zhang Q, Wu G, Richards E, Jia S, Zeng C (2007). Universal primers for HBV genome DNA amplification across subtypes: a case study for designing more effective viral primers. Virol. J. 4:92
Crossref
|
|