Short Communication
ABSTRACT
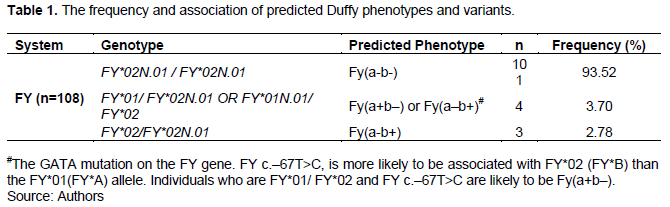
A variant is an alternative nucleotide located at a specific region of a gene. 48 genes encode for human red cell blood group systems. Variants within these genes encode for alleles, which can be highly polymorphic. The blood group gene loci jointly display all types of inherited variants to include single nucleotide variants insertions/deletions and structural variants. In Africa, there is limited information on the red cell variants. The aim of the study was to establish the frequency Duffy red blood cell variants among the donors in Kenya. The study employed next sequencing method, descriptive statistics and results presented in form of a table. The findings show that Duffy system has three variants to include; homozygous for FY*Null GATA regulatory box variant FY*02N.01/FY*02N.01 or Fy(a–b–) found in 93.52% (101/108), while FY*01/FY*02 and FY c.–67T>C predicted Fy(a+b–) or Fy(a–b+) at 3.70% (4/108) while FY*02/FY*02N.01 predicted Fy(a–b+) at 2.78% (3/108). This study recommends an extended research involving large sample size and introduction of extended phenotyping in the identification of FY antigens population.
Key words: Duffy, variant, allele, FY*A, FY*B, molecular, genome, gene.
Abbreviation: EDTA, Ethylenediaminetetraacetic acid; HDFN, haemolytic disease of the fetus and the new born; HTR, haemolytic transfusion reaction; KNBTS, Kenya National Blood Transfusion Service; RBCs, red blood cells; SNVs, single nucleotide variants.INTRODUCTION
A variant is an alternative nucleotide located at a specific region within a gene or genome. A gene variant is a permanent change in the DNA sequence that makes up that genome. 48 genes encode for human red cell blood group systems; variants within these genes encode for alleles, which can be highly polymorphic. The blood group gene loci jointly display all types of inherited variants to include single nucleotide variants (SNVs), insertions/ deletions (indels), and structural variants (Johnsen, 2015). The aim of the study was to establish the frequency Duffy red blood cell variants among the donors in Kenya
The Duffy blood group system is a glycoprotein/cytokine receptor (the binding site for Plasmodium vivax). It is located on chromosome 1q22-q23 and is the first autosomal gene to have ever been revealed in human beings (1950) (Dean, 2005).
Duffy antigens are of clinical significance in transfusion and transplant medicine. This is because they are capable of mounting an immunological response if introduced to those who lack them by forming specific antibodies against (alloimmunisation). Alloimmunisation is mostly associated with those patients who need continuous transfusion as in cases of sickle cell diseases and cancer. The variants and phenotype have also been implicated in minor HDFN and parental immunization in women of child bearing age resulting to HDFN in next deliveries (Dean, 2005).
Duffy blood system is encrypted by FY gene. This gene has two major genotypes (alleles) FY*A and FY*B only differing by one nucleotide (Gly42Asp) and are expressed codominantly (Lopez et al., 2015). The ISBT gene name is FY number 008 Blood Group Alleles, HUGO Gene (ACKR1). The gene is composed of two exons and genomic NM_002036.4 (transcript) FY*01 allele) FY*A or Fya. The alleles form the basis of the four phenotypes to include Fy(a+b-), Fy(a+b+), Fy(a-b+) and Fy(a-b-) (Guelsin et al., 2011; Howes et al., 2011; Dean, 2005).
MATERIALS AND METHODS
A total of 108 anonymous left over blood samples collected in EDTA vacutainers were identified, selected and prepared for molecular genotyping. Manual Genomic DNA extraction was accomplished by the use of QIAamp whole blood DNA mini kit (250 51106) as per manufacturer’s instructions (Qaigen Germany). Library preparation was performed using a custom blood grouping enrichment panel (Roulis et al., 2020), and the Illumina DNA prep Enrichment protocol as per manufacturer’s instructions (Illumina Inc., San Diego, CA, USA).
Inclusion criteria
Samples collected in 4 ml ETDA tubes, non-reactive for transfusion transmission infections (TTIs), with no haemolysis.
Exclusion criteria
This includes all samples that failed the required inclusion criteria in preparation for the manual DNA extraction.
RESULTS
The finding shows that Duffy system has three variants which include; homozygous for FY*Null GATA regulatory box variant FY*02N.01/ FY*02N.01 or Fy(a–b–) found in 101/108 (93.52%), while FY*01/ FY*02 and FY c.–67T/C predicted Fy(a+b–) or Fy(a–b+) found in 4/108 (3.70%) and FY*02/ FY*02N.01 predicted Fy(a–b+) found in 3/108 (2.78%) as shown in Table 1.
DISCUSSION
The finding of this study analysis revealed that Duffy system has two major variants: Fy(a-b-) and Fy(a+b-) or Fy(a-b+), respectively. Fya and b are highly immunogenic proteins and thus capable of mounting an immunological response that results to alloimmunisation. The formed antibodies are implicated in causing hemolytic transfusion reactions, transplant rejection as well as haemolytic disease of the fetus and the new born if introduced to recipients via transfusion, transplantation and pregnancy. Patients who are identified to be alloimmunized such as those who receive multiple transfusions (SCD, oncology, dialysis, post-partum hemorrhage should be given FY antigen negative blood and organs for safer outcomes.
Comparing the results with other studies in different populations, there are some minor variations that were observed. In South Africa, black population Fy(a-b-) 39% while Fya+ Fyb+ was reported to be 2% (Govender et al., 2021). In Saudi Arabia, type Fy(a-b-) was identified in 11.11% (Benahadi et al., 2014). In the Brazilian Japanese population, FY study has shown that FY*01/FY*01(Fya+) occurrence was 59.33%, FY*01/FY*02 (Fya+ Fyb+) was 38.27% and FY*02/FY*02 (Fyb+) was 2.39% (Flôres et al., 2014; Höher et al., 2018).
In sub-Saharan African people, FY null phenotypes frequency has been shown to be ≥95%. In the Indianpopulation, the FY types indicated that Fy (a+b-) was the most common 42.1%, Fy (a+b-) most occurring in the white population 49%, and Fy (a-b-) most prevalent in the black people 68% (Howes et al., 2011). Most of those expressing the null phenotypes were from the black population (Makroo et al., 2013). A study conducted by Rodriguez (2020), revealed that most Africans have Fy(ab-) that is the Fy null phenotype. He also observed that 68% of the African Americans and 88-100% and 90% people of West Africa were Duffy negative Fy(ab-). The null Fy(ab-) type silences the gene in the ertthryroid cells. He together with other authors have shown that this null Duffy phenotype is rare in the Caucasians (Rodriguez, 2020).
CONCLUSION
The study revealed that there are two major variants associated with the Duffy system in the blood donor population: FY*02N.01/FY*02 N.01 97 (89.37%) and FY*01/FY*02N.01OR FY*01N.01/FY*02 11 (10.19%). These variants give rise to two phenotypes or blood types Fy null Fy (ab-) erythroid cells only and Fy (a+b-) or Fy (a-b+), respectively. The results from different studies have shown some variations in different population. However, despite of the differences, the null phenotypes Fy (a-b-) is the most common in the African population.
RECOMMENDATION
The study employed molecular genotyping (NGS) to determine the presence of the red cell variants in the blood donor population. In addition to confirmed presence of red cell variants, valuable knowledge is presented on the basis for improved investigations of the red cell variants in the Kenya. This study outcome is very beneficial in resolving cases associated with adverse transfusion reaction especially in ongoing and massively transfused patients (Sickle cells, cancer cases among others) matching and as variants reference gene bank in Kenya. The results of this study in combination with others related to other blood groups red cell variants will be useful in instituting a blood grouping molecular reference laboratory for Kenya and East Africa community. It will also be useful evidence to consider use of extended phenotyping in blood bank laboratories both in Kenya and East Africa in the identification of rare phenotypes that are of clinical significance in transfusion medicine. More research on this field in Kenya and East Africa will enrich the gene data base and strengthen the blood grouping laboratories in the investigations of complicated complex cases as well as enhance collaborations nationally and internationally.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
ACKNOWLEDGEMENT
The authors thank the Ministry of Health-Kenya, Tissue and Transplant Authority Kenya (formally Kenya National Blood Transfusion Service), Mount Kenya University, Queensland University of Technology Australia & Development, Clinical Services and Research, and Australian Red Cross Blood Service.
ABBREVIATIONS
EDTA, Ethylenediaminetetraacetic acid; HDFN, haemolytic disease of the fetus and the new born; HTR, haemolytic transfusion reaction; KNBTS, Kenya National Blood Transfusion Service; RBCs, red blood cells; SNVs, single nucleotide variants.
REFERENCES
|
Benahadi A, Boulahdid S, Adouani B, Laouina A, Mokhtari A, Soulaymani A, Hajjout K, Benajiba M, Alami R (2014). Mapping rare erythrocyte phenotypes in Morocco: A tool to overcome transfusion challenges. Journal of Blood Transfusion 2014:e707152. |
|
|
Dean L (2005). The Duffy blood group. In Blood Groups and Red Cell Antigens [Internet]. National Center for Biotechnology Information (US). |
|
|
Flôres MR, Visentainer JEL, Guelsin GAS, Fracasso AS, Melo FC, Hashimoto, MN, Sell AM (2014). Rh, Kell, Duffy, Kidd and Diego blood group system polymorphism in Brazilian Japanese descendants. Transfusion and Apheresis Science 50(1):123-128. |
|
|
Govender L, Prakashchandra RD, Pillay P, Jentsch U (2021). Molecular red cell genotyping of rare blood donors in South Africa to enhance rare donor-patient blood matching. African Journal of Laboratory Medicine 10(1):1400. |
|
|
Guelsin GA, Sell AM, Castilho L, Masaki V.L, Melo FC, Hashimoto MN, Hirle, LS, Visentainer JE (2011). Genetic polymorphisms of Rh, Kell, Duffy and Kidd systems in a population from the State of Paraná, southern Brazil. Revista Brasileira de Hematologia e Hemoterapia 33(1):21-25. |
|
|
Höher G, Fiegenbaum M, Almeida S (2018). Molecular basis of the Duffy blood group system. Blood Transfusion 16(1):93-100. Available at: |
|
|
Howes RE, Patil AP, Piel FB, Nyangiri OA, Kabaria CW, Gething PW, Zimmerman PA, Barnadas C, Beall CM, Gebremedhin A, Ménard D, Williams TN, Weatherall DJ, Hay SI (2011). The global distribution of the Duffy blood group. Nature Communications 2(1):1-10. |
|
|
Johnsen JM (2015). Using red blood cell genomics in transfusion medicine. Hematology 2015(1):168-176. |
|
|
Lopez GH, Morrison J, Condon JA, Wilson B, Martin JR, Liew YW, Flower RL, Hyland CA (2015). Duffy blood group phenotype-genotype correlations using high?resolution melting analysis PCR and microarray reveal complex cases including a new null FY*A allele: The role for sequencing in genotyping algorithms. Vox Sanguinis 109(3):296-303. |
|
|
Makroo RN, Bhatia A, Gupta R, Phillip J (2013). Prevalence of Rh, Duffy, Kell, Kidd and MNSs blood group antigens in the Indian blood donor population. The Indian Journal of Medical Research 137(3):521-526. Available at: |
|
|
Rodriguez C (2020). African or African American? That is the question. The Duffy antigen and the Chemokine Storm in Covid-19. Qeios. Available at: https://doi.org/10.32388/ADIHQ5 |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0