ABSTRACT
Sorghum chafer, Pachnoda interrupta (Olivier) is a damaging pest of sorghum, other cereals and horticultural crops in Africa which results in complete crop loss. Currently, the management strategies rely heavily on chemical pesticides, which do not provide effective control. There is evidence showing that microbial biocontrol agents have the potential to control P. interrupta. In this study, the entomopathogenic fungi isolated from soils and insects in Ethiopia were identified using morphological and cultural characteristics. These were then evaluated for viability and virulence against Galleria mellonella (Fabricius) (Lepidoptera, Pyralidae) larvae and P. interrupta adults. Firstly, 116 Metarhizium spp. and Beauveria spp. were considered. The isolates were evaluated based on high viability as measured by percentage germination on SDA media. Only 56 isolates had greater than 70% viability. Secondly, these were further evaluated using conidial suspensions applied to G. mellonella larvae at a concentration of 1×108conidia/ml. Four Beauveria spp. and three Metarhizium spp. isolates which caused over 60% mortality were selected for final virulence assay against adults of P. interrupta. The selected isolates were further confirmed by PCR amplification of ITS4 and ITS5 gene regions and chi1 and chi4 primers. Finally, the seven isolates were evaluated for virulence against P. interrupta at a dose of 1 mg dry conidia/10 beetles under laboratory condition. Mortality of P. interrupta ranged from 14% for isolate 9604 to 82% for isolate PPRC51. Dose-response tests showed that the LD50 of PPRC2 (0.62 mg/10 beetles) and PPRC51 (0.55 mg/10 beetles) isolates were not significantly different from each other. The results demonstrated the high potential of the two isolates (PPRC51 and PPRC2) as microbial biocontrol agents. However, field evaluation of the isolates should be performed for their development into a mycopesticide against P. interrupta.
Key words: Pachnoda interrupta, Metarhizium anisopliae, Beauveria bassiana, bioassay, ITS4, ITS5.
Sorghum chafer, Pachnoda interrupta (Olivier) (Coleoptera: Scarabaeidae) is one of the most destructive polyphagous insect pests of sorghum and over 35 other important crops in Africa in general and in Ethiopia in particular (Grunshaw, 1992; Hiwot, 2000). Some of the crops and host species damaged by the adults of P. interrupta recorded in Ethiopia, Eritrea, Mali, Cameroon, Nigeria, and Somalia include cereals such as sorghum, pearl millet, rice, maize, ornamentals such as roses, vegetables such as cucumber and okra, oil crops such as sunflower, niger seed and sesame; fruits such as guava, banana, mango and papaya and other tree plants such as Acacia spp. (Andemeskel, 1987; Grunshaw, 1992; Troure and Yehouienou, 1995; Jago, 1995; Ratnadas and Ajayi, 1995; MOA and EARO, 1999; Sastawa and Lale, 2000; Hiwot, 2000). P. interrupta out-breaks cover wide geographic areas (MOA and EARO, 1999; Yeraswork, 2000; Asmare and Yeshitila, 2014) and completely destroy sorghum fields at the milk stage (Tsedeke, 1988). Grain abortion and panicle sterility caused by the pest can result in 100% yield loss even on insecticide treated sorghum fields (Yitbarek and Hiwot, 2000; Yeraswork, 2000).
Control of P. interrupta mainly depends on the use of insecticides (Seneshaw, 2001) which incurs high costs to small farmers and entails environmental, human and animal health related hazards. Moreover, controlling adult beetles through application of insecticides on scattered sorghum fields does not provide long-lasting control because of continuous re-infestation (Seneshaw and Mulugeta, 2002; Yitbarek, 2008). Thus, alternative control methods that can contribute to integrated management of P. interrupta need to be sought. One of the possible methods is to develop efficient biological control agents that can control larvae and adults at the breeding sites (Seneshaw and Mulugeta, 2002). Biological control agents, which include naturally occurring microbial bio-control agents, have great potential in controlling pest populations with little or no detrimental effects on human health and the environment (Khan et al., 2012). Among them, the entomopathogenic fungi (EPF) are most preferred because of ease of production and application, mode of action that does not need ingestion of the entomopathogen by the target pest (Butt, 2002; Wang and St. Leger, 2007; Thomas and Read, 2007). EPF are especially important when used within IPM programs as they are compatible with pesticides (Lacey and Goettel, 1995; Wraight et al., 2007). These fungi have also restricted host ranges and thus cause little or no harm to non-target organisms such as parasitoids and predators (Goettel and Hajek, 2001; Vestergaard et al., 2003, Hajek and Goettel, 2007).
Developing microbial bio-control agents in modern scientific studies requires identification of selected isolates. Characterization and analysis of genetic traits of fungi and other micro-organisms is facilitated by accurate and powerful molecular tools (Inglis et al., 2012). Moreover, molecular identification of potential EPF is gaining acceptance as an important first step for successful development of myco-insecticides (Islam et al., 2014). Particularly, molecular tools such as PCR based analysis of DNA are used as standard procedures for identification and phylogenetic comparisons between EPF (Jensen et al., 2001; Destefano et al., 2004) and as molecular markers for species identification (Driver et al., 2000; Entz et al., 2005; Islam et al., 2014). The objective of this study was, therefore, to identify and evaluate the potentials of native entomopathogenic fungi for the development of myco-insecticides against P. interrupta.
Isolation and sources of EPF
All the EPF were isolated from soils and infected insects. The source institutions, places of collection, habitats/ host genera and germination of isolates used for the experiments are presented in Table 1.
Screening against G. mellonella
Initial identification
The key for the identification of major genera of fungi described by Humber (2005) was used to initially identify all the 116 isolates. Only those isolates identified as belonging to the genera Beauveria or Metarhizum were screened for viability and pathogenecity against larvae of G. mellonella.
Screening of isolates
Screening for viability
All isolates initially identified as belonging to either Beauveria or Metarhizium (101 isolates), were subjected to germination test to select the ones with reasonable viability. Spores were harvested from the surfaces of media using sterile metal spatula and added to test tubes containing 10 ml of sterilized Tween 80 (0.01%) to make a stock suspension. The concentration of the stock suspension was adjusted to 3 × 106 conidia/ml using an improved neubaour heamocytometer and 100 µl of the suspension was then spread plated on SDA media in 90 mm diameter Petri-dishes. After 24 h of incubation at 25°C, a sterile cover slip was put on each Petri-dish and percent germination was determined by counting at least 300 conidia under a compound microscope at 400 X magnification. A conidium was declared germinated if it showed a growth as big as its size. Three replicate Petri-dishes were used for each isolate.
The isolates selected from screening for viability were further screened for pathogenicity against G. mellonella larvae. To obtain larvae, adult moths were collected in 500 ml flasks containing folded tissue paper impregnated with water and honey. When eggs were laid, the tissue paper was removed from the flasks and put in plastic rearing boxes containing 180, 50 and 180 g of honey, wheat bran and glycerol, respectively as feed for the larvae. The boxes were incubated in the dark at 20°C for four weeks. The resulting fourth to fifth instar larvae were used for the bioassays.
Stock suspensions were prepared from the respective isolates as for the initial screening and spore concentration was adjusted to 1 × 108 conidia/ml. The Metarhizium spp. isolates were bio-assayed in two separate assays, while the Beauveria spp. isolates were bio-assayed in three separate assays. For each isolate, 10 larvae were immersed in 10 ml conidial suspension for 10 s in a sterile beaker after which the contents of the beaker were passed through a sterile muslin cloth. The larvae were then transferred into 55 mm diameter sterile plastic Petri-dishes containing filter paper and incubated at room temperature (22-26°C). The control was treated with 10 ml solution of Tween 80(0.01%). A completely randomized design (CRD) with four replications was used for the experiments.
Mortality of the larvae was assessed every day for 10 days. Dead larvae were surface sterilized by briefly immersing in 70% of alcohol and quickly rinsing with sterile distilled water twice. Finally the larvae were transferred to a sterile Petri-dish containing wet filter paper, sealed with parafilm and incubated at room temperature to check for mycosis.
Screening against P. interrupta
Three Metarhizium anisopliae and four Beauveria bassiana isolates which caused 60% or more larval mortality were selected from the screening against G. mellonella for final bio-assay against P. interrupta. Adult beetles were collected during the mating season of 2013 from breeding areas around Mendubo village (10°50'N, 040°05'E; altitude 1206 m.a.s.l.) in Oromia zone of Amhara Regional State in Ethiopia and kept in plastic baskets containing moistened sterile soil collected from the same area. The baskets had side openings for aeration and the tops were covered with muslin cloth to prevent beetles from escaping. Collected beetles were fed with slices of ripe banana and observed for any natural infection for 10 days before being used for bio-assays.
Conidia of the selected isolates were re-isolated from sporulating cadavers of G. mellonella larvae and grown on SDA media at 25°C. After 2 to 3 weeks of incubation, conidia were harvested with sterile metal spatula and collected in sterile Petri-dishes. To remove excess moisture, the conidia were oven-dried at 30°C overnight before the bioassays. The number of spores/mg of the isolates were 3.03 × 108 , 2.9 × 108, 3.5 × 108, 2.9 × 108 , 2.0 × 108, 1.4 × 108, 1.7 × 108 and 3.9 × 108 for isolates 9604, 9609, MP3POST, Melke36, DLCO131, Green Muscle, PPRC51 and PPRC2, respectively. Ten beetles were put in sterile 300 ml plastic tubs with perforated lids. One miligram spores of each isolate were applied on top of the beetles in each of the plastic tubs and the beetles were allowed to move in the tubs for 30 min. The beetles were then transferred to 120 mm diameter plastic Petri-dishes containing moist filter paper and incubated at room temperature for 10 days. During incubation, beetles were fed with slices of ripe banana changed every other day. To provide adequate moisture, 1 ml of sterile distilled water was added to the Petri-dishes every day. Mortality was assessed every day and dead beetles were removed and surface sterilized with 70% ethanol and rinsed thrice in sterile distilled water. The surface sterilized beetles were then transferred to sterile Petri-dishes containing moist filter paper and incubated at 25°C to check for sporulation and to confirm death due to fungal infection. The experimental design was a completely randomized design (CRD) with four replications. The control was treated with the respective spores killed at 80°C in an oven for 48 h. The bioassay was repeated after 75 days using beetles from the October 2013 population from Rassa area (09° 57’ N and 040° 04’ E) and freshly sub-cultured and harvested spores as in the first bio-assay. Mean percentage mortality from the two experiments were used as measures of virulence. Comparison among selected most virulent isolates was done using their respective LT50 and LT90 values. The commercialized myco-insecticide Green Muscle containing M. anisopliae spores as an active ingredient was used as a standard in the two bioassays.
Dose-response test
A dose-response bioassay was conducted on the three promising isolates (PPRC51, PPRC2 and MP3POST) obtained from the screening against P. interrupta. To determine the range of conidial doses used for the test, initial bioassay experiments were conducted with six different doses (0.05, 0.1, 0.25, 0.5, 0.75 and 1 mg/10 beetles). The two lower doses (0.05 and 0.1 mg/10 beetles) did not result in any mortality within 10 days and therefore, excluded from the dose range. Thus, only four doses were used for the final dose-response tests using probit analysis: Finney (1964). Dry conidia were prepared for the evaluation against P. interrupta and weighed on a sensitive balance (Adventure ™ USA). All the procedures used for the screening against P. interrupta were repeated exactly except that four different doses were applied.
Molecular characterization
The characterization of the selected seven isolates obtained from the screening against P. interrupta was done at molecular level using the internal transcribed spacer (ITS) gene region (ITS4 and ITS5) and Chitinase (chi1 and chi4) primers.
DNA extractions
EPF isolates from Ethiopia were cultured on (SDA) media. For each fungal isolate, 50 to 100 mg of the mycelia was scrapped and DNA were extracted using the Isolate II Plant DNA Kit (Bioline), following the manufacturer’s protocol. The resultant DNA was eluted in 50 μL of the elution buffer and stored at -20ºC, until further processing. The extracted DNA quality was then checked using a Nanodrop 2000/2000c Spectrophotometer.
PCR amplification of the ITS4 and ITS5 gene region
Amplifications were carried out for the rDNA region of the fungal isolates using the ITS primers (White et al., 1990). The PCR was carried out in a total volume of 30 μL containing 0.2 μM of each primer (ITS 5; 5’GGA AGT- AAA- AGT- CGT- AAC- AAG -G 3’ and ITS 4; 5’ TCC- TCC- GCT -TAT -TGA –TAT- GC 3’, respectively), 5X My Taq Reaction Buffer (Bioline), 1.25 mM MgCl2, 1 unit My Taq DNA polymerase (Bioline) and 3 μL of genomic DNA template. Typical cycle conditions were as follows: Initial denaturation at 95°C for 1 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 59°C for 40 s, and primer elongation at 72°C for 1 min followed by a final extension at 72°C for 10 min done in an Arktik programmed thermal cycler.
PCR amplification using chi 1 and chi 4 primers
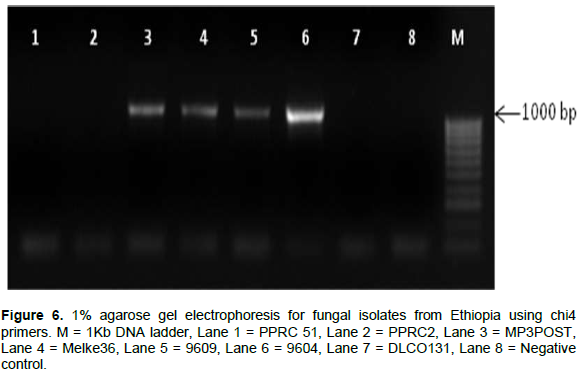
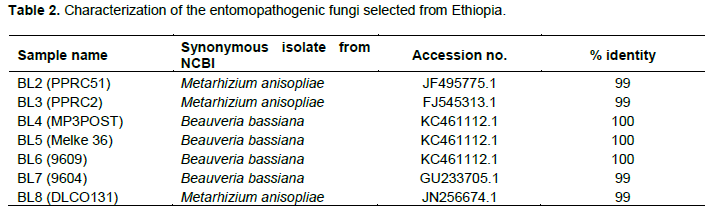
The results from the ITS gene region amplification were also confirmed by amplifying the chitinase gene region using redesigned chi 1 and chi 4 primers. In this regard, markers were designed from the chitinase gene to discriminate between the two species through gel electrophoresis. The PCR was carried out in a total volume of 30 μl containing 0.2 μM of each specific primer, 5X My Taq Reaction Buffer (Bioline), 1 unit My Taq DNA polymerase (Bioline) and 3 μL of DNA template. Chi 1 primer set gave a target region of 800 bp, while the Chi 4 primer set had a target size of approximately 1000 bp. Typical cycle conditions were as follows: Initial denaturation at 95°C for 1 min, followed by 35 cycles of denaturation at 95°C for 15 s, specific annealing for 15 s, and primer elongation at 72°C for 10 s followed by a final extension at 72°C for 10 min that was done in an Arktik programmed thermal cycler.
Detection and analysis of the PCR products
The amplified PCR products were resolved through a 1% agarose gel stained with ethidiumbromide (10 mg/ml) and subjected to electrophoresis set at 70 volts for 1 h (Bio-Rad model 200/2-0 power supply and wide mini-sub cell GT horizontal 56 electrophoresis system, Bio-Rad laboratories, Inc., USA), followed by visualization of the DNA under UV-illumination. The gel photo was analyzed and documented using the KETA GL imaging system from Wealtec Corp.
Gel extraction and purifications
All successfully amplified PCR products for each of the targeted gene regions were excised and purified using the Isolate II PCR and Gel Kit from Bioline following the manufacturer’s instructions. A total of 17 purified PCR products were sent to Macrogen Inc, Europe Laboratories, the Netherlands for bi-directional sequencing.
Sequencing data analysis
This was done using bioinformatics tools and software. The sequences obtained were edited by Chromas Lite version 2.1.1 software and the consensus sequences from both the forward and reverse strands generated. For conclusive identification, the consensus reads generated were queried through BLASTN, at the GenBank data base hosted by the National Centre of Biotechnology Information USA (NCBI). This was also to check for similarity with organisms already identified. Furthermore, the consensus sequences generated were multi-aligned using Clustal X (version 2.1). The multiple alignments created were used to generate a phylogenetic tree by use of Mega software (version 6.06).
Statistical analysis
Mortality data were corrected using Abbot’s formula (Abbott, 1925), arcsine transformed and subjected to the ANOVA procedure of SAS version 9.0. Percentage viability data were also analyzed in the same way. Means were separated using LSD and Tukey’s Honestly Significant Difference (HSD) for screening experiments against G. mellonella and P. interrupta, respectively. LT50, LT90 and LD50, values were estimated with probit analysis for correlated data followed by ANOVA and mean separation with LSD. Probit analyses were done using SPSS version 17.
Initial identification
Out of 116 isolates collected from the different sources, 101 isolates were preliminarily identified using morpho-logical and cultural characteristics as Metarhizium spp. or Beauveria spp. Fifteen of the isolates were identified as not belonging to the two genera.
Screening of isolates
Viability of isolates
The germination percent of the 101 isolates evaluated for viability ranged from 0 to 99 (data not shown). The isolates with greater than 70% germination were considered sufficiently viable and selected for further screening (56 isolates) against G. mellonella (Table 1).
Effects of fungal isolates on G. mellonella
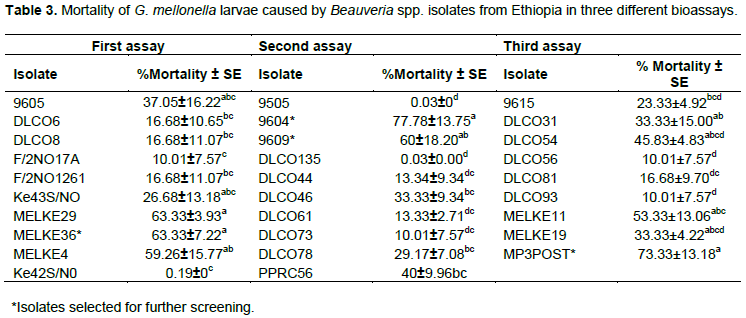
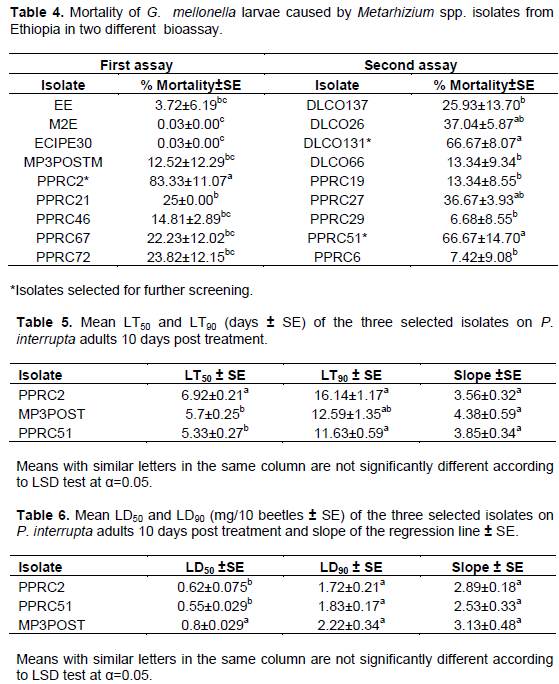
Corrected percent mortality of G. mellonella larvae due to fungal isolates significantly varied among isolates of each of Beauveria spp. and Metarhizium spp. (Tables 3 and 4). The Beauveria spp. isolates caused between 0 and 63%, 0 and 78%, and 0 and 73% mortality in the first, second and third bioassays, respectively. Similarly, the Metarhizium spp. isolates caused 0 to 83% and 0 to 67% mortality in the first and second bioassays, respectively.
Effects of fungal isolates on P. interrupta
There was a highly significant variation among the isolates (P = 0.0003, df = 7, 16, F = 8.01) in causing mortality to P. interrupta (Figure 2). The lowest mean percent mortality was caused by the B. bassiana isolate 9604 (14.08%) which was not significantly different from B. bassiana isolate 9609 (27.97%), Melke36 (15.75%), M. anisopliae isolates DLCO131 (25%) and Green Muscle (20.38%). The highest mortality to P. interrupta was caused by PPRC51 (82.40%) which did not significantly differ from the B. bassiana isolate MP3POST and the M. anisopliae isolate PPRC2 which caused 79.63 and 77.14% mortality, respectively. Based on the results of the virulence assays, two M. anisopliae isolates (PPRC51 and PPRC2) and one B. bassiana isolate (MP3POST) which caused over 75% mortality to P. interrupta adults were selected. The time taken by the three selected isolates to cause death to 50 and 90% of the experimental insects (LT50 and LT90 days) is shown in Table 4. The LT50 (days) of the selected isolates varied significantly (P = 0.0087, df = 2, 6, F = 11.6) with the lowest (5.33 days) recorded from PPRC51 followed by MP3POST (5.7 days). The highest (6.92 days) was recorded due to PPRC2 which was significantly different from both isolates. However, there was no significant difference (P = 0.057, df = 2, 6, F = 4. 78) in the LT90 (days) of the three isolates. Figure 1 shows growth and sporulation of PPRC51 and PPRC2 isolates on P. interrupta adults during the bio-assays.
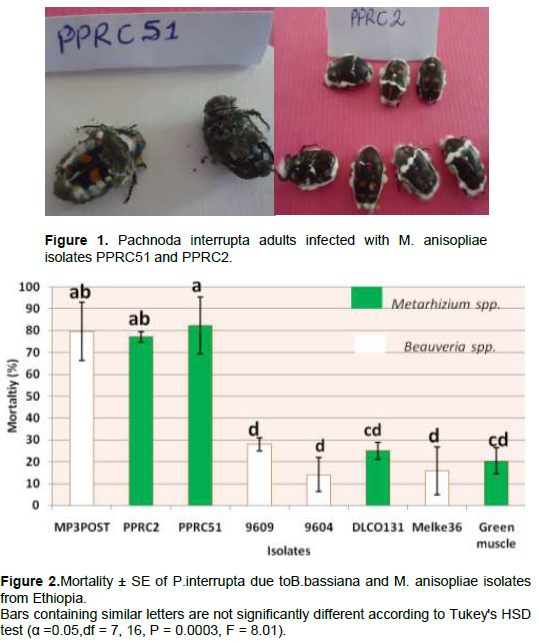
Dose-response test
Mortality of P. interrupta adults at different doses of the selected M. anisopliae and B. bassiana isolates is shown in Figure 3. There were no significant mortality differences within each dose except for the dose of 0.5 mg in which the B. bassiana isolate MP3POST showed significantly lower mortality (P = 0.039, F = 5.78, df = 2 , 6 ) than the two M. anisopliae isolates. The mean LD50 of the three isolates also varied significantly (P = 0.025, F = 7.12, df = 2,6) among the isolates with isolate MP3POST requiring higher dose (0.8 mg/10 beetles) to kill 50% of the test insects than PPRC2 (0.62 mg/10 beetles) and PPRC51 (0.55 mg/10 beetles) isolates, which were not significantly different from each other (Table 5). There were no significant differences in the mean LD90 among all the three isolates (P = 0.41, F = 1.04, df = 2, 6).
Molecular identification
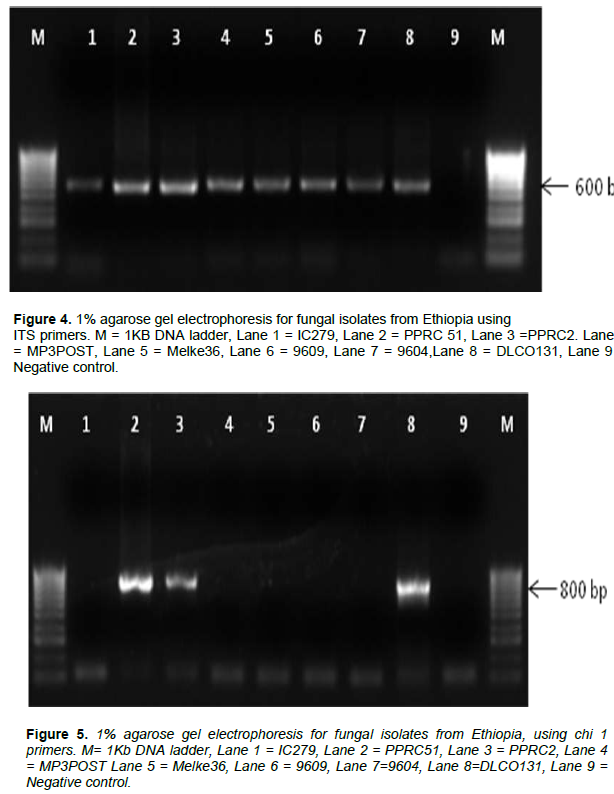
The internal transcribed spacer region (ITS) was successfully amplified in all of the isolates as shown in Figure 4. Figures 5 and 6 show the amplification of the two fungal genera Metarhizium and Beauveria, respectively using chi1 and chi4 genes. The BLAST searches corresponded to sequences registered under M. anisopliae and B. bassiana and were provided with accession numbers as depicted in Table 2 which confirms the identities of the 7 isolates. Three of the samples were linked to M. anisopliae and four to B. bassiana in the GenBank database.
The sequences from the ITS gene region were used to generate a phylogenetic tree (Figure 7). The evolutionary history was inferred using the Neighbor joining method (Saitou and Nei, 1987). The optimal tree with the sum of branch length = 0.19117607 is shown in Figure 4. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method (Kimura, 1980) and are in the units of the number of base substitutions per site. The analysis involved 7 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Non coding. All positions containing gaps and missing data were eliminated. There were a total of 518 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013). The tree clustered the Metarhizium and Beauveria species into two clusters, as expected.

Fungi can be identified using morphological charac-teristics but this method is not adequate due to ambiguous descriptions and limited availability of morphological keys (Fernandes et al., 2010). However, the method can be used to minimize time and resource expenses when dealing with large number of isolates as a preliminary identification tool as employed in this study. The finally selected 7 entomopathogenic fungi from Ethiopia screened against P. interrupta, were identified as M. anisopliae and B. bassiana by the PCR amplification of the ITS regions of rDNA and chitinase genes. The ITS has been widely and effectively used to amplify DNA from a wide range of fungi (Kendall and Rygiewicz, 2005). This region is highly conserved and is commonly used as a sole tool or supplemented with other universal sequences (e.g. tubulin and actin) for identification, characterization and phylogenetic analysis of fungal isolates (Balazy et al., 2008). The ITS region has been used in other studies for successful identification of entomopathogenic fungi (Islam et al., 2014). The results from amplification of the ITS region from the current study were confirmed with amplification with the chitinase genes. This functional gene confirmed the identity of the Ethiopian isolates as belonging to M. anisopliae and B. bassiana, as expected. Similarly, Chitinase genes have been used in other studies as molecular markers for M. anisopliae identification and characterisation (Bogo et al., 1998; Kang et al., 1999; Enkerli et al., 2009).
Merid et al. (2016) have recently reported the use of Metarhizium spp. for field control of P. interrupta which showed up to 71% mortality within 15 days of infection. Researches on other scarab beetles (Lacey et al., 1994; Klein and Lacey, 1999; Cuthbertson et al., 2012) have also shown the potential of these fungi for control of coleopterous insects. This current study has demon-strated the importance of these entomopathogenic fungi as potential microbial biocontrol agents for P. interrupta. Among the seven isolates, the M. anisopliae isolate PPRC51 caused 82.40 % mean mortality on field collected P. interrupta adults within 10 days post application and had the shortest LT50 (5.33 days). The B. bassiana isolate MP3POST and M. anisopliae isolate PPRC2 also caused 79.67 and 77.14% mortality, respectively. In a similar study, Lacey et al. (1994) observed 100% mortality of the Japanese beetle, P. japonica adults within 8 and 9 days after application of M. anisopliae and B. bassiana dry conidia, respectively, using similar doses Gindin et al. (2006) also reported 85% mortality of the red palm weevil, Rhynchophorus ferrugineus, two weeks after contact with dry conidia of M. anisopliae indicating the potential of the fungi for biocontrol of coleopterous insects. Although isolate MP3POST needed significantly higher LD50 than PPRC2 and PPRC51, its LT50 was not significantly different from that of PPRC51.
The findings of this study have indicated the potential use of the indigenous EPF isolates against P. interrupta. The isolates PPRC51, PPRC2 and MP3POST are as found the potential candidates for development of myco- insecticide against P. interrupta as a component of an integrated management strategy of the pest. However, field studies using appropriate formulation under high insect population conditions and more research on mass production characteristics and shelf life of the isolates are needed.
The authors have not declared any conflict of interests.
Special thanks to Dr. Sunday Ekesi who invited the first author to the International Center of Insect Physiology and Ecology (ICIPE) at Nairobi, Kenya and covered the laboratory expenses. Complements also go to the staff of arthropod pathology unit of ICIPE particularly to Mr. Levy Odhiambo for the molecular laboratory work. The authors also thank Mr. Tesfaye Hailu and Mis. Aberash Challa of Ambo Plant Protection Center for the collaborations. The Addis Ababa University is acknowledged for providing the necessary funds for the author’s visit to ICIPE where part of this work was done. The bio-innovate project is also acknowledged for the financial support to this work. Dr. Mattias C. Larsson was financed by the Linnaeus initiative IC-E3, supported by Formas and the Swedish Science Council.
REFERENCES
|
Abbott WS (1925). A Method of Computing the Effectiveness of an Insecticide. J. Econ. Entomol. 18:265-267.
Crossref
|
|
|
|
Andemeskel W (1987). Handbook of insect pests of major crops in Eritrea Administrative Region, and their control. Asmara University. P 132.
|
|
|
|
|
Asmare D, Yeshitila M (2014). Ecology and field biology of the sorghum chafer, Pachnoda interrupta (Olivier) (Coleoptera: Scarabaeidae) in Ethiopia. Adv. Entomol. 2:8-13.
Crossref
|
|
|
|
|
Balazy S, Wrzosek M, Sosnowska D, Tkaczuk C, Muszewska A (2008). Laboratory trials to infect insects and nematodes by some acaropathogenic Hirsutella strains (Mycota: Clavicipitaceous anamorphs). J. Invertebr. Pathol. 97:103-113.
Crossref
|
|
|
|
|
Bogo MR, Rota CA, Pinto Jr. (1998). A chitinase encoding gene (chit1 gene) from the entomopathogen Metarhizium anisopliae: isolation and characterization of genomic and full length cDNA. Curr. Microbiol. 37(4):221-225.
Crossref
|
|
|
|
|
Butt TM (2002). Use of entomogenous fungi for the control of insect pests, In. Esser K, Bennett JW (eds). Mycota, Springer, Berlin, pp. 111-134.
Crossref
|
|
|
|
|
Cuthbertson AGS, Mathers JJ, Blackburn LF, Powell ME, Marris G, Pietravalle S, Brown MA, Budge GE (2012). Screening commercially available biocontrol agents for the control of Aethina tumida (Coleoptera: Nitidulidae) in the UK. Insects 3:719-726.
Crossref
|
|
|
|
|
Destefano RHR, Destefano SAL, Messias CL (2004). Detection of Metarhizium anisopliae within infected sugarcane borer Diatraea saccharalis (Lepidoptera, Pyralidae) using specific primers. Genet. Mol. Biol. 27:245-252.
Crossref
|
|
|
|
|
Driver F, Milner RJ, Trueman JWH (2000). A taxonomic revision of Metarhizium based on a phylogenetic analysis of rDNA sequence data. Mycol. Res. 104:134-150.
Crossref
|
|
|
|
|
Enkerli J, Ghormade C, Oulevey C, Widmer F (2009). PCR RFLP analysis of Chitinase genes enables efficient phenotyping of Metarhizium aninopliae var. anisopliae. J. Invertebr. Pathol. 102(2):185-188.
Crossref
|
|
|
|
|
Entz SC, Johnson DL, Kawchuk LM (2005). Development of a PCR-based diagnostic assay for the specific detection of the entomopathogenic fungus Metarhizium anisopliae var. acridum. Mycol. Res. 109:1302-1312.
Crossref
|
|
|
|
|
Felsenstein J (1985). Confidence limits on phylogenies: An approach using the boot-strap. Evolution 39:783-791.
Crossref
|
|
|
|
|
Fernandes E KK, Keyser CA, Chong JP, Rangel DEN, Miller MP, Roberts DW (2010). Characterization of Metarhizium species and varieties based on molecular analysis, heat tolerance and cold activity. J. Appl. Microbiol. 108:115-128.
Crossref
|
|
|
|
|
Finney DJ (1964). Probit analysis: a statistical treatment of the sigmoid response curve. Cambridge: University Press, 318p.
|
|
|
|
|
Gindin G, Levski S, Glazer I, Soroker V (2006). Evaluation of the entomopathogenic fungi Metarhizium anisopliae and Beauveria bassiana against the red palm weevil Rhynchophorus ferrugineus. Phytoparasitica 34(4):70-379.
Crossref
|
|
|
|
|
Goettel MS, Hajek AE (2001). Evaluation ofnontarget effects of pathogens used for management of arthropods, In. Wajnberg, E. Scotland, J.K. Quimby, P.C, (Eds.), Evaluating Indirect Ecological Effects of Biological Control. CABI Press, Wallingford, UK. pp. 81-97.
|
|
|
|
|
Grunshaw JP (1992). Field studies on the biology and economic importance of Pachnoda interrupta (Coleoptera: Scarabaeidae) in Mali West Africa. Bull. Ent. Res. 82:19-27.
Crossref
|
|
|
|
|
Hajek AE, Goettel MS (2007). Guidelines for evaluating effects of entomopathoges on non-target organisms. In. Lacey LA, Kaya HK (eds). Field Manual of Techniques in Invertebrate Pathology: Application and Evaluation of Pathogens for Control of Insects and other Invertebrate Pests. Springer Publishing, Dordrecht, The Netherlands, Pp. 815-833.
Crossref
|
|
|
|
|
Hiwot L (2000). Historical background on the pest status and control of sorghum chafer, Pachnoda interrupta (Coleoptera: Scarabaeidae) in Ethiopia. In. Proceedings of the workshop on the development of monitoring and controlstrategy against sorghum chafer, Pachnoda interrupta (Coleoptera: Scarabaeidae) in Ethiopia, February – 2 March, 2000, MOA, Addis Ababa. Pp. 9-15. 28.
|
|
|
|
|
Humber RA (2005). Entomopathogenic Fungal Identification USDA-ARS Plant Protection Research Unit US Plant, Soil & Nutrition Laboratory Tower Road Ithaca, NY 14853-2901. of Techniques in Invertebrate Pathology (2nd edition). Academic Press, New York, USA.
|
|
|
|
|
Inglis GD, Enkerli J, Goettel MS (2012). Laboratory techniques used for entomopathogenic fungi: Hypocreales. In: Lacey, L. A. (ed.). Manual
Crossref
|
|
|
|
|
Islam MT, Omar D, Shabanimofrad M (2014). Molecular identification and virulence of six isolates of Metarhizium anisopliae (Deuteromycotina: Hyphomycetes) to Bemisia tabaci Q biotype. J. Asia Pac. Entomol. 17:237-241.
Crossref
|
|
|
|
|
Jago ND (1995). Population monitoring and crop loss assessment in integrated pest management of panicle pests of sorghum and pearl millet. In: Nwanze, K.F, Youm, O. (eds). Proceedings of an international consultative workshop on panicle insect pests of sorghum and pearl millet, 4-7 October, 1993, ICRISAT Sahelian Centre, Niamey, Niger. Andhra Pradesh, India: ICRISAT. Pp. 103-113.
|
|
|
|
|
Jensen AB, Thomsen L, Eilenberg J (2001). Intra-specific variation and host specificity of Entomphthora uscae sensu strict isolates revealed by random amplified polymorphic DNA, universal primed PCR, PCR- restriction fragment length polymorphism, and conidial morphology. J. invertebr. Pathol. 78:251-259.
Crossref
|
|
|
|
|
Kang SC, Park S, Lee DG (1999). Purification and characterization of a novel chitinase from entomopathogenic fungus Metarhizium anisopliae. J. Invertebr. Pathol. 73(3):273-281.
Crossref
|
|
|
|
|
Kendall JM, Rygiewicz PT (2005). Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 5:28.
Crossref
|
|
|
|
|
Khan S, Guo L, Maimaiti Y, Mijit M, Qiu D (2012). Entomopathogenic fungi as microbial biocontrol agents. Mol. Plant Breed. 3:63-79.
Crossref
|
|
|
|
|
Kimura M (1980). A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120.
Crossref
|
|
|
|
|
Klein MG, Lacey L (1999). An attractant trap for autodissemination of entomopathogenic fungi into populations of the Japanese beetle Popillia japonica (Coleoptera: Scarabaeidae). Biocontrol Sci. Technol. 9(2):151-158.
Crossref
|
|
|
|
|
Lacey LA, Goettel MS (1995). Current developments in microbial control of insect pests and prospects for the early 21st century. Entomophaga 40:3-27.
Crossref
|
|
|
|
|
Lacey LA, Martins A, Ribeiro C (1994) The pathogenicity of Metarhizium anisopliae and Beauveria bassiana for adults of the Japanese beetle, Popillia japonica (Coleoptera: Scarabeidae). Eur. J. Entomol. 3:313-319.
|
|
|
|
|
Merid NG, Tibebe DB,Yitbarek W, Jonas MB, Ylva H, Emiru S (2016). Metarhizium sp. isolated from dead Pachnoda interrupta (Coleoptera:Scarabaeidae) as a potential entomopathogenic fungus for the pest insect: proof-of-concept for autodissemination. Int. J. Trop. Insect Sci. 36(1):1-9.
Crossref
|
|
|
|
|
MOA, EARO (1999). The significance, distribution and control of sorghum chafer, Pachnoda interrupta (Olivier) Coleoptera: Scarabaeidae in Amhara and Afar regions. 10-13 March, 1999, MOA and EARO, Addis Ababa, Ethiopia. 71 p.
|
|
|
|
|
Ratnadas A, Ajayi O (1995). Panicle Insect pests of sorghum in West Africa. In. Nwanze KF, Youm O (eds). Panicle Insect Pests of Sorghum and Pearl Millet: Proceedings of an International Consultative Workshop, 4-7 Oct. 1993, ICRISAT Sahalian Center, Niamey, Niger. Patanchera 502 324, Andhra Pradesh, India: International Crops Research for Semi-Arid Tropics. Pp. 29-38-113.
|
|
|
|
|
Saitou N, Nei M (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425.
|
|
|
|
|
Sastawa BM, Lale NES (2000). Efficacy of host plant resistance, sowing date modification and intercropping as methods for the control of Pachnoda interrupta (Olivier) in pearl millet in the Nigerian Sudan savanna. J. Arid Environ. 46:249-262.
Crossref
|
|
|
|
|
Seneshaw A (2001). Activity report on insect pest management with fungi: A mass production technique for farmers. Cooperative development research project C-16-125. Ambo PPRC, Ethiopia.
|
|
|
|
|
Seneshaw A, Mulugeta N (2002). Study on the Biology of sorghum chafer, P. Interrupta (Coleoptera: Scarabaeidae) under laboratory condition. Pest Manage. J. Ethiop. 6:31-36.
|
|
|
|
|
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30:2725-2729.
Crossref
|
|
|
|
|
Thomas MB, Read AF (2007). Can fungal biopesticides control malaria? Nature reviews. Microbiology 5:377-383.
Crossref
|
|
|
|
|
Troure K, Yehouenou A (1995). Les insects de I'epi de mil en Afrique de I'Ouest. In: Nwanze KF, Youm O (eds). Proceeding of an International Consultative Workshop on Panicle Insect Pests of Sorghum and Pearl Millet, 4-7 October, 1993 ICRISAT Sahelian Centre, Niamey, Niger. Andhra Pradesh, India: ICRISAT.
|
|
|
|
|
Tsedeke A (1988). Insect and mite pests of horticultural and miscellaneous plants in Ethiopia. IAR Hand Book No. 1, Institute of Agricultural Research. Addis Ababa. 115 p.
|
|
|
|
|
Vestergaard S, Cherry A, Keller S, Goettel M (2003). Hyphomycete fungi as microbial control agents Chapter 3. In: Hokkanen HMT, Hajek AE (eds.). Environmental Impacts of Microbial Insecticides. Kluwer Academic Publishers, Dordrecht, The Netherlands. pp. 35-62.
Crossref
|
|
|
|
|
Wang C, St. Leger RJ (2007). The Metarhizium anisopliae perillipin holog MPL1 regulates lipid metabolism, appressoral turgor pressure and virulence. J. Biol. Chem. 282:21110-21115.
Crossref
|
|
|
|
|
White TJ, Bruns T, Lee S, Taylor JW (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. and White, T.J. (Eds.). PCR Protocols: A Guide to Methods and Applications. Academic Press Inc. New York. Pp. 315-322.
Crossref
|
|
|
|
|
Wraight SP, Inglis GD, Goettel S (2007). Fungi. In: Lacy, L.A. and Kaya, H.K. (eds.). Field Manual of Techniques in Invertebrate Pathology (2nd edition.). Application and evaluation of pathogens for control of insects and other invertebrate pests. Section IV-4. Springer, Dordrech, The Netherlands.
|
|
|
|
|
Yeraswork Y (2000). The importance, distribution and current status of Sorghum chafer, Pachnoda interrupta (Olivier) in Amhara Region In. Proceedings of The Workshop on the Development on Monitoring and Control Strategy Against sorghum shafer, Pachnoda interrupta (Olivier), (Coleoptera: Scarabaeidae) in Ethiopia. Addis Ababa. pp. 24-35.
|
|
|
|
|
Yitbarek W (2008). Electrophysiological and behavioral responses of sorghum chafer, pachnoda interrupta (coleoptera:scarabaeidae) to host plant volatile compounds. PhD. Dessertation. Addis Ababa University.
|
|
|
|
|
Yitbarek W, Hiwot L (2000). Preliminary yield loss assessment on sorghum due to sorghum chafer, Pachnoda interrupta (Olivier) in Amhara Region. In Proceedings of the Workshop on the Development, Monitoring and control strategy against sorghum chafer, Pachnoda interrupta (Olivier), (Coleoptera: Scarabaeidae) in Ethiopia. Addis Ababa. pp. 39-43.
|
|