Full Length Research Paper
ABSTRACT
Fusarium wilt is one of the most devastating diseases in sesame production in Uganda, caused by a fungus called Fusarium oxysporum f.sp. sesami. Its incidence ranges from 17.1 to 73.3%. Some sesame genotypes have been reported to resist Fusarium wilt; however, have not been precisely determined. In this study, 30 sesame genotypes that included released varieties, improved elite breeding lines and introductions were screened in the screenhouse under high pathogen pressure following artificial infection using five isolates of F. oxysporum f.sp sesami. The results revealed that sesame genotypes showed different response to the pathogen and thus disease development among the genotypes. The genotype effect was significant (P≤0.001) for disease incidence. Two genotypes (EM15-1-5 and Sesim 2) were identified and rated moderately resistant to Fusarium wilt (37.3 and 33.8%), respectively. Whereas, seven genotypes showed moderate susceptibility and 21 genotypes were susceptible for Fusarium wilt infection. No genotype was identified as being immune to the disease. It is noteworthy for sesame breeding programme in Uganda to continue evaluating other genotypes from existing germplasm which were not tested for resistance to Fusarium wilt in this study. Also more germplasm should be assembled and screened for resistance to the disease and other agronomic traits.
Key words: Fusarium oxysporum f. sp. sesame, incidence, resistance.
INTRODUCTION
Sesame is an important food and cash crop in Uganda, providing livelihoods for many households. However, its yield is very low, often below 1000 kg/ha. Low sesame yield in Uganda is due to some abiotic and biotic constraints. Abiotic constraints include fluctuations of rainfall and temperature, poor soil management while biotic factors include pests and diseases. Fusarium wilt (Fusarium oxysporum f. sp. sesami) is one of the diseases devastating sesame production in Uganda. Its incidence had been reported to range between 17.1 and 73.3% (Egonyu et al., 2005). Elsewhere the disease had been reported to cause yield loss ranging from 50 to 100% (El-Bramawy et al., 2009). F. oxysporum f. sp. sesami is a soil borne, parasitic pathogen which interacts with host plant (Bayoumi and EL-Bramawy, 2007). When inside the plant the pathogen feeds on plant’s nutrients and colonizes the roots cells which then spread to other parts of the plants through the water transporting vessels (xylem). The pathogen grows and produce mycelia which leads to blockage of water supply to the plant consequently the plants develops wilt symptoms (Elewa et al., 2011; Joshi, 2018). Fusarium wilt in Uganda is managed by using cultural practices such as early planting, intercropping, crop rotation, and burning the crop residues. These cultural practices however are not efficient in managing the disease. Early planting is intended to enable the crop to escape from being affected by Fusarium wilt pathogen which is severe toward the end of the rainy season. This practice unfortunately exposes the crop to waterlogging and other diseases such as leaf spot (Egonyu et al., 2005). Intercropping and crop rotation are meant to reduce the pathogen population in soil and thus reduce disease incidence and severity. The effectiveness of these two methods is also reduced due to the effective survival strategies of the pathogen (Okungbowa and Shittu, 2012). Developing and growing disease resistant varieties is thus the most effective, environmental friendly, less costly and long term solution to disease management in crops (Shabana et al., 2014). There is therefore need to identify or develop sesame varieties with resistance to Fusarium wilt. Sesame genotypes such as Sesim 1 and Sesim 2 have been developed in Uganda and are reported to be resistant to Fusarium wilt (Anyanga-Okello et al., 2016b). Resistance levels in these genotypes however, have not been precisely determined. There is also need to increase the sesame germplasm base with resistance to Fusarium wilt to facilitate resistance breeding. This study therefore, was done to screen sesame genotypes for resistance to F. oxysporum f.sp. sesami.
MATERIALS AND METHODS
Pathogen isolation and identification of Fusarium species associated with sesame wilt
Fusarium spp. isolates were obtained from wilted sesame plants collected from a field at the National Semi-Arid Resources Research Institute (NaSARRI), Eastern Uganda. Fields at NaSARRI are known to be the hot spot of Fusarium wilt. Wilted plant samples (Figure 2) were uprooted gently; roots and the portion of the stems exhibiting reddish-brown necrosis (like crown rot), cut and placed in paper envelopes. The samples were taken to the Biotechnology Laboratory at Makerere University, Agriculture Research Institute, Kabanyolo (MUARIK) to isolate the disease causing organism (Figure 1).
Isolation of the pathogen
Pathogen isolation was carried out according to Kavak and Boydak (2006) with some modifications. The roots and stem samples were washed under running tap water and air dried in a laminar flow hood. The air dried stems were cut in small pieces approximately 5 mm long and dissected longitudinally to observe internal vascular discoloration. Stem pieces that exhibited internal vascular discoloration and roots were further cleaned using distilled water and disinfected using 2.5% v/v commercial bleach (JIK) for 5 min followed by 70% ethyl alcohol for 1 min. Disinfected pieces were then rinsed three times in sterile distilled water and blotted dry using a sterile napkin in a Laminar flow hood.
The sterilized stems and roots were chopped into small pieces and placed on PDA media in separate Petri dishes and incubated at 27°C under alternating 12 h light and dark conditions for seven days. Cultures were observed for conidia production. Those with conidia were transferred to fresh PDA media and incubated at same temperature and light conditions for another seven days. Pure cultures were then made from single hyphal tips, again on PDA. Mature cultures were observed macro and microscopically. Macroscopically cultures were observed for growth morphology and pigmentation of colonies (Burgess et al., 1994; Leslie and Summerell, 2006). The colony morphology, growth habit and pigmentation were observed and recorded.
For microscopic identification, mycelia from each isolates were teased on a microscope slide and mounted on a microscope to observe presence as well as the shape of macro and micro conidia and chlamydospores including their number of septa (Leslie and Summerell, 2006). Microscopically, many of the cultures conformed to Fusarium spp. five isolates (SEFU 1-SEFU 5) were consequently selected for use in this study.
Molecular identification of Fusarium spp. associated with sesame wilt from cultured isolates
Molecular identification of the fungal isolates was done based on DNA sequencing of the translation elongation factor-1 alpha (TEF-1 alpha) and Internal Transcribed Spencer regions (ITS). For this purpose, fungal DNA was extracted and subjected to polymerase chain reaction (PCR) as described by Namasaka et al. (2017). The TEF-1α was amplified using forward (TEF-Fu3f: Bioneer corporation 5’-GGT ATC GAC AAG CGA ACC AT-3) and reverse (TEF-Fu3r: Bioneer Corporation 5’- TAG TAG CGG GGA GTC TCG AA- 3’) primers while the ITS was amplified using the forward and reverse primers ITS5: 5’-GGAAGTAAAAGTCGTAACAAGG-3’ and ITS4 KY0: 5’-TCCTCCGCTTWTTGWTWTGC-3’), respectively. DNA amplification was carried out in an ARKTIK Thermocycler (IngabaBiotechTM, Model: 5020). From each of the amplified sample, 5 µl were subjected to gel electrophoresis on 1.5% agarose gel pre-stained with Gel-red fluorescent dye (Botium) in 1x TBE buffer at a 130 V for 30 min alongside 100 bp DNA ladder. Gel documentation was done using the Benchtop UV Trans-illuminator (BioDoc-ItTM Imaging System, 8.0’’ LCD/LM-20, PlN 97-0165-02, 230V-50Hz) and the bands of each sample scored by comparing its position with a specific band of the ladder in the gel to the ladder chart provided (Bioneer Corporation). Five isolates that conformed to F. oxysporum f. sp. sesame based on microscopic, macroscopic and DNA sequence analysis were selected and subjected to pathogenicity tests using 3 susceptible varieties before being used to screen germplasm.
Screening of sesame germplasm against Fusarium wilt pathogen
Thirty (30) sesame genotypes were sourced from NaSARRI (Table 1) and screened for Fusarium wilt resistance. The genotypes comprised local materials, improved lines and introductions from Tanzania (Table 1). These genotypes were screened two times (1st screening and 2rd screening). Screening was conducted in a screen house at MUARIK which is located in Central Uganda (32° 37’E, 0° 28’N) at an altitude of 1200 m above sea level. MUARIK receives an average annual rainfall of 1200 mm with a daily temperature ranging from 17 to 33°C (Namasaka et al., 2017).
Experimental design, inoculum production and inoculation
The test genotypes were grown in a chamber constructed inside a screenhouse. The chamber was constructed from a white transparent polythene sheet to increase temperature and humidity for pathogen multiplication (Namasaka et al., 2017). Five pathogen isolates were used to screen the thirty genotypes. The isolates were first cultured on sterilized sorghum in the laboratory for 21 days at room temperature. The fully colonized sorghum seed was then used to inoculate the sterilized soil in plastic pots. Inoculation was done using 75 g of inoculum for every 2 kg of soil. Inoculated pots were then arranged in a split plot design with two replicates. Fungal isolates were the main plot treatment while genotypes were sub-plot treatments. Planting was done three days after inoculation. A total of 15 seeds per genotype were planted in each pot. Pots were watered regularly to ensure proper plant growth. Screening of this 30 genotypes was done two times (experiment 1 and 2) (Table 7) in the screenhouse.
Data collection
Phenotypic observations were made daily for Fusarium wilt symptoms from emergence to physiological maturity of the crop. The experimental germplasm was observed for necrotic lesions on the crown, yellowing of lower leaves, downward turning of growing plant (epinasty), wilting and death. Data were also collected for plant stand per pot and number of diseased plants per genotype. Plant stand was recorded two weeks after emergence and used to determine emergence percentage (EM %) and seed rotten%. Number of diseased plants per genotype was recorded at an interval of ten days from seedling to physiological maturity and used to compute disease incidence (DI %).
Data analysis
Data were subjected to Analysis of Variance (ANOVA) in Genstat 18th Edition Software. Fisher Protected Least Significant Difference (LSD) test at 5% probability level was used to compare treatment means. Genotypes were then grouped according to their resistance levels using the scale developed by Kavak and Boydak (2006) with some modifications (Table 2). The following are equations used to compute EM and DI%.
where  is the observation value for genotype ith, jth and kth,
is the observation value for genotype ith, jth and kth,  is mean,
is mean, 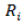 is the replication effect for ith ,
is the replication effect for ith ,  is the isolate effect for jth ,
is the isolate effect for jth ,  is the replication interacting genotype effect for the ith and jth,
is the replication interacting genotype effect for the ith and jth,  is the genotypes effect for kth,
is the genotypes effect for kth,  is the isolate interacting genotype for kth and jth, and
is the isolate interacting genotype for kth and jth, and 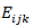 is the experimental error effect.
is the experimental error effect.
RESULTS
Isolation and identity of the Fusarium spp. associated with sesame wilt
Based on phenotypic characteristics and DNA sequence data, five isolates were identified and belonged to F. oxysporum. All five isolates produced white to magenta or magenta-pink cotton like mycelia on PDA (Figure 2) but in dark conditions the colour changed to and remained purple. The isolates produced macroconidia, microconidia and chlamydospores. Macroconidia were moon shaped and multiseptate (Figure 3). These features are associated with Fusarium spp. DNA sequences of the translation elongation factor-1 alpha (TEF-1 alpha) and Internal Transcribed Spencer regions (ITS) revealed 99% congruence with several F. oxysporum accessions deposited in data bases (Tables 3 and 4). All the five isolates resulted into a wilt on the susceptible genotypes, proving that they all were F. oxysporium f.sp. sesami.
Response of 30 genotypes against five isolates of F. oxysporum f. sp. sesami
In Table 5, the isolate effect was non-significant in the first screening but significant (P≤0.05) during the second screening. The performances of genotypes were significant (P≤0.001) for all traits measured both in the first and second screening. Interaction between genotype and isolate was only significant (P≤0.001) for DI% during the first screening. A significant effect of the interaction of genotypes by isolates was noted for all traits during the second screening.
For genotypes response during the first and second screening, the analysis indicated that 22 and 25 genotypes, respectively had EM% above 70% (Tables 6 and 7). Across experiments, 24 genotypes had EM% above 70% (Table 8). During the first screening (Table 6), genotype (Sesim2//5181)-2-2-1 (86.0%) recorded the highest EM% and ranked first although the difference was not significant from 6 other genotypes. The genotype with the lowest EM% was Mtwara 2009 (31.4%). However, this was not significantly different from two genotypes Lindi 2002 (34.4%) and Naliendele 92 (44.3%). The highest seed rot percentage was recorded in the genotype Lindi 2002 (94.3%) and was significantly higher than values recorded on the other 29 genotypes. The lowest seeds rot percentage was recorded in genotype Ajimo A1-6//7029-1-1 (14.3%) and was significantly lower than seven other genotypes. For DI%,(Sesim 2//5181)//UCR3-1-19-2 (95.5%) recorded the highest value and it was significantly different from 24 other genotypes. Genotype EM15-1-5 (31.4%) recorded the lowest DI%. This was however not significantly different from two genotypes Renner 1-3-1-17 (31.5%) and Sesam2 (32.4%).
Contrary to the first screening, in the second screening (Table 7), the highest EM% was recorded in the genotype Mtondo 2015 (97.0%). This was significantly higher than 26 other genotypes. Genotype Renner 1-3-1-17 (48.0%) recorded the lowest EM% but differences were not significant from 18 other genotypes. The genotype with the highest seed rot percentage was Renner 1-3-1-17 (52.0%). However, this was not significantly different from nine other genotypes. Lowest seed rot percentage was recorded in the genotype Mtondo 2015 (3.0%). Genotype Mtwara 2009 (90.5%) recorded the highest DI%. The lowest DI% was recorded in a genotype Local 158-5 (34.3%). This figure was not significantly different from 28 other genotypes.
Genotypes were not consistent with respect to resistance to Fusarium wilt across the two experiments. During first screening, three genotypes (EM15-1-5, Renner 1-3-1-17 and Sesim 2) were categorized as moderately resistant, four genotypes were moderately susceptible, and 17 genotypes were susceptible while six genotypes were highly susceptible (Table 6). Results of the second screening showed that three genotypes (Local 158-5, Sesim 2 and Local 158//6022) were moderately resistant, 12 genotypes were moderately susceptible, 14 were susceptible and only one genotype (Mtwara 2009) was categorized as highly susceptible (Table 7). Across experiments, only two genotypes (Sesim 2 and EM15-1-5) were moderately resistant, seven were moderately susceptible while the majority were categorized as susceptible (Table 8).
While performance for isolates during the first screening showed that their effect was not significant for all traits recorded (Figure 4), in the second screening (Figure 5) the effect was significant. In the second screening, the highest EM% was recorded in isolate SEFU3 (85.1%) and it was not significant from other two isolate SEFU4 (81.5%) and SEFU2 (81.4%). Lowest EM% was recorded in isolate SEFU1 (72.6%) and was significantly different from three other isolates except SEFU5 (77.2%). Highest seed rot percentage was recorded from in isolate SEFU1 (28.5%) but was not significantly different from isolate SEFU5 (22.8%). The lowest seed rot percentage was in isolate SEFU5 (15.7%) and was not significantly different from three other isolates. Highest DI% was recorded from isolate SEFU5 (64.7%) although this was not significantly different from isolate SEFU1 (61.8%), SEFU2 (64.6%) and SEFU4 (63.1%). Isolate SEFU3 resulted in the least in DI% (38.3%).
DISCUSSION
Thirty sesame genotypes were screened for resistance to F. oxysporum f. sp. sesame in screenhouse conditions A range of symptoms were observed on the inoculated crop. These consisted of seed rot, reddish-browning of hypocotyls, damping off, reddish-brown necrotic lesions on stem and roots, wilting and plants death.
The findings demonstrated that the performance of all 30 sesame genotypes was significantly different in all the measured traits. This implies that genotypes were genetically diverse, presenting an opportunity for finding useful genotypes in the screened set. During the two screenings, most of the genotypes had percentage emergence values above 70% implying that the majority of the genotypes were able to germinate. This study has shown that F. oxysporum. f. sp. sesami does not result into serious seed rot that affects germination rate. This is not unusual since typical wilt pathogens do not cause much seed rots.
The study also showed that there was variation in disease incidence among the genotypes tested. During the first screening, three genotypes (EM15-1-5, Renner 1-3-1-17 and Sesim 2) had disease incidence below 40% of these genotypes; however, only Sesim 2 scored below 40% incidence in the second screening. Interestingly, two other genotypes (that is, Local 158-5 and Local 158//6022) also had disease incidence levels below 40%. This means that some genotypes were not consistent in their resistance level suggesting that resistance of Fusarium wilt in sesame may also be influenced by other factors in addition to genetic ones. These factors such as carbon dioxide, temperature, texture, soil pH, and moisture in most cases are environmental and have been reported to interact with soil borne pathogens in a way that influences disease occurrence. The more carbon dioxide or less pH in soil, the more multiplication of fungi (Anonymous, 1988; Stover, 1958; Tyagi and Paudel, 2014) unexpectedly the plant fail to fight against the infestation. Moreover, Agrios (2005) pointed out that when the temperature is not favorable for the pathogen may trigger the plants to escape.
The study showed a significant genotype isolate effect for disease incidence. This suggested that some pathogen isolates reacted differently with different sesame genotypes. This may be due to differences in then genetic make-up of either sesame genotypes or the pathogen isolates. Other environmental factors could also have contributed towards the observed differences. None of the genotype was immune or resistant to the disease, majority being susceptible. Other studies have also failed to find sesame genotypes immune to Fusarium wilt (El- Bramawy et al., 2001; El-Shazly et al., 1999; Kavak and Boydak, 2006). Sesim 2 was moderately resistant across the two experiments. Anyanga et al. (2016a) had earlier reported that in addition to Sesim 1, Sesim 2 was resistant to Fusarium wilt. However, this study found Sesim 1 to be susceptible.
The isolate effect was significant for EM%, Seed rot% and DI%. This indicated that isolates were different in aggressiveness towards the genotypes tested. Isolate SEFU1, SEFU2, SEFU4 and SEFU5 were more aggressive compared to SEFU3. Therefore, any of these four isolates (SEFU1, SEFU2, SEFU4 and SEFU5) can be used to screen other sesame germplasm for wilt resistance. However, performance of these isolates differed in the two experiments probably due to differences in soil pathogen population. Several factors influence survival and saprophytic multiplication of fungi in the soil (Agrios, 2005; Stover, 1958; Tyagi and Paudel, 2014). Differences in these factors may have influenced performance of these isolates.
CONCLUSION AND RECOMMENDATION
The findings of this study indicate that there are sesame genotypes that are moderately resistant to F. oxysporum f. sp. sesami in Uganda. Two genotypes (EM15-1-5 and Sesim 2) were moderately resistant to F. oxysporum f. sp. sesami. These genotypes can be used for commercial production as well as breeding activities. The majority of genotypes including Sesim 1 were susceptible to the pathogen however those with good attributes can be improved for resistance to wilt. It is significant for sesame breeding programme in Uganda to continue evaluating other genotypes from existing germplasm which were not tested for resistance to Fusarium wilt in this study. Also, for a long term solution to wilt, more germplasm should be assembled and screened for resistance to the disease, other agronomic traits and reaction to biotic stresses.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the Alliance for a Green Revolution in Africa (AGRA) for the full financial support of the study and Makerere University Agricultural Research Institute, Kabanyolo (MUARIK) for allowing the research to be conducted in their screenhouse. They also thank National Semi-Arid Resources Research Institute (NaSARRI) administration for giving them tested genotypes and field to conduct this study.
REFERENCES
|
Agrios GN (2005). Plant Pathology, 5th ed. Elsevier Academic Press, United State of America. |
|
|
Anonymous (1988). Report on plant disease. University of Illinois at Urbana-Champaign. |
|
|
Anyanga WO, Rubaihayo P, Gibson P, Okori P (2016a). Combining ability and gene action in sesame (Sesamum indicum L) elite genotypes by diallel mating design. Journal Plant Breeding Crop Science 8:250-256. |
|
|
Anyanga-Okello W, Rubaihayo P, Gibson P, Okori P (2016b). Genotype by environment interaction in sesame (Sesamum indicum L.) cultivars in Uganda. African Journal Plant Science 10:189-202. |
|
|
Bayoumi TY, EL-Bramawy MAS (2007). Genetic analyses of some quantitative characters and Fusarium wilt disease resistance in sesame. African Crop Science Social 8:2198-2204. |
|
|
Burgess LW, Summerell BA, Bullock S, Gott KP, Backhouse D (1994). Laboratory manual for Fusarium research, 3rd ed. University of Sydney. |
|
|
Egonyu JP, Kyamanywa S, Anyanga W, Ssekabembe CK (2005). Review of pests and diseases of sesame in Uganda. African Crop Science Conference Process 7:1411-1416. |
|
|
El-Bramawy MA, Veverka K, Veverka S, El-Shazly MS, El-Sattar MA, El-Ashary MA, Ammar SE (2001). Evaluation of resistance to Fusarium oxysporum f.sp. sesami in hybrid lines of sesame (Sesamum indicum L.) under greenhouse conditions. Plant Protection Science 37:74-79. |
|
|
El-Bramawy MAEHS, El-Hendawy SE, Shaban WI (2009). Assessing the suitability of morphological and phenotypical traits to screen sesame accessions for resistance to Fusarium wilt and charcoal rot diseases. Plant Protect Science 45:49-58. |
|
|
Elewa IS, Mostafa MH, Sahab AF, Ziedan EH (2011). Direct effect of biocontrol agents on wilt and root-rot diseases of sesame. Achieve Phytopathology Plant Protect 44:493-504. |
|
|
El-Shazly MS, Wahid OAA, EL-Ashry MA, EL-Barmawy MA (1999). Evaluation of resistance to Fusarium wilt disease in sesame germplasm. International Journal Pest Management 45:207-210. |
|
|
Joshi R (2018). A review of Fusarium oxysporum on its plant interaction and industrial use. Journal of Medicinal Plants Research 6:112-115. |
|
|
Kavak H, Boydak E (2006). Screening of the resistance levels of 26 sesame breeding lines to Fusarium wilt disease. Plant Pathology Journal 5:157-160. |
|
|
Leslie JF, Summerell BA (2006). The Fusarium laboratory manual, First edit. ed. Blackwell Publishing, 2121 State Avenue, Ames, Iowa 50014, USA. |
|
|
Namasaka RW, Tusiime G, Orawu M, Gibson P, Nyiramugisha J, Edema R (2017). Evaluation of Cowpea Genotypes for Resistance to Fusarium redolens in Uganda. America Journal Plant Science 8:2296-2314. |
|
|
Okungbowa FI, Shittu HO (2012). Fusarium wilts: An overview. Environment Resource Journal 6:83-102. |
|
|
Shabana R, Abd El-mohsen AA, Khalifa MMA, Saber AA (2014). Quantification of resistance of F 6 sesame elite lines against Charcoal-rot and Fusarium wilt diseases. Advance Agriculture Biology 1:144-150. |
|
|
Stover RH (1958). Some factors influencing survival and saprophytic multiplication of F. oxysporum f. sp. cubense in soil. Canada Journal Botany 36:311-324. |
|
|
Tyagi S, Paudel R (2014). Effect of different pH on the growth and sporulation of Fusarium oxysporum : The causal organism of wilt disease of Tomato. International Journal Basic Applied Biology 2:103-106. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0