ABSTRACT
In August 2012, vegetable crops including potato, cabbage, bell pepper and carrot showing symptoms of maceration and water soaked lesions on their tuber, leaf and fruit were collected from major vegetable growing areas in north-west of Iran. Physiological and biochemical assays divided the isolates into two main groups according to their ability to grow at 37°C. In addition, these two groups were differentiated by some biochemical characteristics such as utilization of α –methyl glucoside and maltose, reducing substrates from sucrose and gelatin liquefaction. Partial sequence of polymerase chain reaction (PCR) product from reaction of putative Pectobacterium spp. with 16S rRNA confirmed the results obtained from physiological and biochemical assays used for identification of the bacterium. All of the causal organisms isolated from infected tissues were identified as Pectobacterium spp. based on biochemical characteristics and PCR amplification of the 16S rRNA gene with specific primers PCCSSF and PCCSSR. Twenty of the isolates were identified as Pectobacterium carotovorum subsp. carotovorum using a polyphasic approach including physiological, biochemical and molecular characteristics. Only six isolates from infected bell peppers and five isolates from infected potatoes identified as Pectobacterium wasabiae and Pectobacterium atrosepticum respectively. Our findings based on physiological, biochemical and molecular assays indicated the occurrence of P. wasabiae as a novel species in bell pepper in Iran. Moreover, genetic diversity of Pcc strains in this study was also performed based on Repetitive extragenic palindromic-PCR (rep-PCR). High genetic variability was revealed among these Pcc strains by rep-PCR typing using two primers sets for ERIC-PCR and BOX-PCR.
Key words: 16S rRNA, repetitive extragenic palindromic- polymerase chain reaction (rep-PCR), Pectobacterium, soft rot, vegetable.
Bacterial soft-rot caused by Pectobacterium spp. has been considered as one of the most recurrent diseases observed in variety of vegetable crops and fruits species worldwide and cause great economic loss of crops (Yahiaoui-Zaidi et al., 2003; Agrios, 2005; Farrar et al., 2000; Toth et al., 2001). Soft-rot symptoms begin as a small water-soaked lesion, which enlarges rapidly in diameter and in depth.
The affected area becomes soft and mushy while its surface becomes discolored and somewhat depressed. Tissues within the affected region become cream colored and slimy, disintegrating into a mushy mass of disorganized plant cells and bacteria. The outer surface may remain intact while the entire contents have changed to a turbid liquid; alternatively, cracks develop and the slimy mass exudes to the surface and, in air, turns tan, gray, or dark brown. A whole fruit or tuber may be converted into a soft, watery, decayed mass within 3 to 5 days. Infected fruits and tubers of many plants are almost odorless until they collapse, and then secondary bacteria grow on the decomposing tissues and produce a foul odor (Agrios, 2005). These groups of bacteria are not specific to plant hosts and produce pectolytic enzymes which damage fleshy fruits, vegetables, ornamental plants and some other agricultural crops, especially in storage (Fahy and Persely, 1983). Hence, characterization of Pectobacterium species and subspecies is very important for effective managing of them (Toth et al., 2001).
The recent studies have clarified the taxonomy of genus and species of Entrobacteriaceae via using biochemical and molecular methods (Gardan et al., 2003). Before these studies, the soft rot causing bacteria of Entrobacteriaceae belonged to Erwinia genus, but the results of recent investigations were led to distinction of Dickeya and Pectobacterium genera (Gardan et al., 2003; Samson et al., 2005). For the first, the Erwinia genus was described in 1917 and it included all of Entrobacteriaceae family that caused the disease in different plants (Perombelon, 1990). However, further studying on diversity of these bacteria especially comparison of 16s rDNA sequence, led to identification of new genera including Dickeya, Brenneria, Entrobacter, Pectobacterium, Pantoea and Samsonia.
The genus Pectobacterium has been divided into several species and subspecies on the basis of molecular, biochemical and host range differences (Gardan et al., 2003; Duarte et al., 2004; Ma et al., 2007; Van der Merwe et al., 2010). To date, five Pectobacterium species have been described: Pectobacterium atrosepticum, Pectobacterium betavasculorum, Pectobacterium carotovorum, Pectobacterium wasabiae (Gardan et al., 2003) and the recent one Pectobacterium brasiliensis (Daurte et al., 2004; Van der Merwe et al., 2010), which is phylogenetically further apart from the other four Pectobacterium spp. (Ma et al., 2007). The species P. betavasculorum and P. atrosepticum constitute an exception to the broad-host range nature of Pectobacterium spp, since they have been reported exclusively on sugar beet and potato respectively (Ma et al., 2007). Whereas the species P. carotovorum, P. brasiliensis, and P. wasabiae have been reported to cause disease on potato and other plant species (Kim et al., 2009). P. carotovorum subsp. carotovorum (Pcc) has a broad host range in comparison with other subspecies of this bacterium and disperses in the subtropical and moderate zones (Perombelon and Kelman, 1980). P. c. subsp. carotovorum has been previously isolated from potato producing areas in Iran (Soltani-Nejad et al., 2005; Firoz et al., 2007; Baghaee et al., 2011). P. atrosepticum potato soft rot and blackleg causing often disperses in the cold and moderate zones (Perombelon and Van der Wolf 2002). However, there are some reports that P. atrosepticum can cause soft rot disease in tomato (Gardan et al., 2003). P. atrosepticum also has been reported in south-west and north of Iran (Zohour Paralak et al., 2007; Baghaee et al., 2011). P. wasabiae previously had been isolated from radish (Goto and Matsumoto, 1987) and recently it has been reported from potato (Kim et al., 2009; Baghaee et al., 2011). To this date, there is no report of P. wasabiae on bell pepper from Iran.
The occurrence of pectolytic enterobacteria causing soft rot have been previously reported in different plants such as radish (Goto and Matsumoto, 1987), aglaonema (Chao et al., 2006), dieffenbachia (MacFadden, 1961; Nieves-Brun et al., 1985), tulip (Boyraz et al., 2006), syngonium (Alippi and Lopez, 2009), potato, tomato, carrot and cabbage (Maisuria and Nerurkar, 2013), artichoke (Loreti et al., 2001), sweet potato, chinese cabbage and eggplant (Golkhandan et al., 2013). The key objective of this research was to identify Pectobacterium spp. as the causal organisms of soft rot through physiological, biochemical and molecular techniques in some vegetable crops in north-west of Iran.
Bacterial strains
The bacterial strains used in this study were isolated from bell pepper (Capsicum annuum), potato (Solanum tuberosum), carrot (Daucus carota) and cabbage (Brassica oleracea) collected from different vegetable growing regions in north-west of Iran which were affected by soft rot disease (Table 1). The disease is characterized by dark and small water-soaked lesions or soft rot symptoms on potato tuber, carrot root, cabbage leaf and bell pepper fruit. The type strains of P. carotovorum subsp. carotovorum (IBSBF-863 = ATCC15713), P. atrosepticum (IBSBF-1819 = ATCC33260), P. betavasculorum (IBSBF-787 = ATCC43762), P. carotovorum subsp. odoriferum (IBSBF-1814 = ICMP11533) obtained from the Instituto Biológico Seção de Bacteriologia Fitopatologia (IBSBF) and reference strain of P. wasabiae (SCRI 488) obtained from Scottish Crop Research Institute (SCRI) were included for comparison in different tests.
Media and cultural condition
Isolation of bacteria from infected samples was carried out according to Perombelon and Van der Wolf (2002). Briefly, after washing the diseased plants small amounts of root, leaf, fruit and tuber samples from the margin of healthy and diseased tissue were homogenized in 1 to 2 drops of sterile water, allowing 2 to 30 min to stand. The suspensions were placed on nutrient agar sucrose (NAS) and eosin methylene blue (EMB) (Schaad et al., 2001). After 24 h of incubation at 25°C, single colonies with white to creamy color and irregular margin on NAS or emerald green on EMB were purified on nutrient agar sucrose medium. All selected isolations were stored in sterile water at 4°C for further investigations.
Physiological and biochemical characterizations
Isolates which were characterized as positive in pectolytic activity were submitted for the biochemical and physiological tests including gram reaction, fermentative metabolism (Hugh and Leifson, 1953), oxidase and catalase activity (Schaad et al., 2001). Hydrolysis of gelatin, casein and lecithin were tested on gelatin agar, skimmed-milk agar and egg-yolk agar respectively (Dickey and Kelman, 1988). They were checked for production of phosphatase, ability to grow at 37°C in nutrient broth, growth in 4 and 5% sodium chloride on nutrient agar at 28°C, urease activity, levan production and gelatin liquefaction (Schaad et al., 2001).
Moreover, production of reducing substances from sucrose, malonate utilization and indole production from tryptophan, anaerobic degradation of arginine and utilization of citrate were examined using previously described methods (Gallois et al., 1992; Gardan et al., 2003).
In addition, the utilization of carbon sources were tested on the basal medium of Ayers et al. (1919) supplemented with 0.1% carbohydrates including lactose, D-fructose, D-Sucrose, D-glucose, D-galactose, trehalose, α-methyl glucoside, D- arabitol, D-melibiose, mannitol, raffinose, sorbitol, maltose, cellubiose and starch. Hypersensitivity reaction (HR) assays were performed with all the isolates (Bauer et al., 1994). Sensitivity assays of the isolates to erythromycin were performed using previously described methods (Schaad et al., 2001; Klement et al., 1990).
Pathogenicity assays
The pathogenicity assay was performed with different vegetable crops including cabbage (Brassica oleracea), potato (Solanum tuberosum), bell pepper (Capsicum annuum), and carrot (Daucus carota). The surface of the potato tubers, bell pepper fruits and carrot roots were sanitized in 70% ethyl alcohol for 30 s and were washed with sterilized distilled water. Tubers and fruits then needle punctured with 10 μl (108 CFU/ml) overnight culture of each strain grown on nutrient broth (NB) for 24 h at 27°C. Inoculated and non-inoculated (control) tubers and fruits were incubated in a moist chamber with 80 to 90% relative humidity at 27°C. After 72 h, macerated tissue was scooped from the tubers and fruits and weighed to determine the extent of tissue maceration (Yap et al., 2004). Pathogenicity tests were carried out similarly on cabbage leaves. However, in this case, the leaves of each plant were separated before sanitization, injected with 10 μl of a bacterial suspension (108 CFU/ml) with a syringe, and then incubated at 28°C for 48 h in a moist chamber with 80 to 90% relative humidity (Kim et al., 2007). Distilled water was used as a negative control. Re-isolations from inoculated plants were confirmed by pectolytic assay, morphology and biochemical tests.
Molecular characterization
Bacterial DNA extraction
Total genomic DNA was extracted from the bacterial isolates according to Cheng and Jiang (2006) protocol. Briefly, 1 ml cell suspension was centrifuged at 8000 g for 2 min. After removing the supernatant, the cells were washed with 400 μl STE Buffer (100 mM NaCl, 10 mM Tris/ HCl, 1 mM EDTA, pH 8.0) twice. Then the cells were centrifuged at 8000 g for 2 min. The pellets were re-suspended in 200 μl TE buffer (10 mM Tris/HCl, 1 mM EDTA, pH 8.0). Then 100 μl Tris-saturated phenol (pH 8.0) was added to these tubes, followed by a vortex-mixing step of 60 s to lyse cells. The samples were subsequently centrifuged at 13000 g for 5 min at 4°C to separate the aqueous phase from the organic phase. 160 μl upper aqueous phases was transferred to a clean 1.5 ml tube. 40 μl TE buffer was added to make 200 μl and mixed with 100 μl chloroform and centrifuged for 5 min at 13 000 g at 4°C. Lysate was purified by chloroform extraction until a white interface was no longer present; this procedure might have to be repeated two to three times. 160 μl upper aqueous phase was transferred to a clean 1.5 ml tube. 40 μl TE and 5 μl RNase (at 10 mg/ml) were added and incubated at 37 C for 10 min to digest RNA. Then 100 μl chloroform was added to the tube, mixed well and centrifuged for 5 min at 13 000 g at 4°C. 150 μl upper aqueous phase was transferred to a clean 1.5 ml tube. The aqueous phase contained purified DNA and was directly used for the subsequent experiments or stored at-20°C.
16S rRNA gene sequence analysis
For a rapid and simple identification and differentiation of Iranian isolates, polymerase chain reaction (PCR) assays were applied with primers PCCSSF (5'-ATAACTACTGGAAACGGTA-3') and PCCSSR (5'-TTCTCTTTGTATACGCCATT-3'). The 16S rRNA gene assays for the isolates were carried out using the method described by Yanagi and Yamasato (1993). PCR was performed in 25 μl of a reaction mixture containing 2.5 μl of 10xPCR buffer, 2.5 mM MgCl2, 0.2 mM of deoxynucleoside triphosphates, 1.5 U of Taq polymerase, 10 pmol of each primer and 3 μl of template DNA. PCR amplification was carried out using thermal cycler (Techne-TC-512) with the following thermal regime; initial denaturing for 3 min at 95°C, 35 cycles of denaturing at 94°C for 60 s, followed by annealing at 49°C for 60 s, elongation at 72°C for 60 s and final extension step at 72°C for 10 min. Amplified DNA fragments were detected by electrophoresis in a 1.0% agarose gel stained with ethidium bromide. The PCR products of representative strains were purified and sequenced by Macrogen Inc. (Seoul, South Korea) using an ABI3730 XL automatic DNA sequencer and the primers PCCSSF and PCCSSR. The identification of the isolates was performed using BLAST (http://blast.ncbi.nlm.nih.gov/blast/Blast.cgi) in NCBI.
Rep-PCR based DNA finger printing
Two repetitive extragenic palindromic- polymerase chain reaction (rep-PCR), genomic fingerprinting methods were performed using single oligonucleotide primer BOX A1R (5'-CTACGGCAAGGCGACGCTGACG-3') and oligonucleotide primer pair ERIC1R (5'-ATGTAAGCTCCTGGGGATTCAC-3') and ERIC2 (5'- AAGTAAGTGACTGGGGTGAGCG-3') based on recommended methods with a little changes (Versalovic et al., 1991; Gillings and Holly, 1997; Weingart and Volksch, 1997). PCR amplifications were performed in 25 μl of a reaction mixture containing 2.5 μl of 10×PCR buffer, 2 mM MgCl2, 0.2 mM of deoxynucleoside triphosphates, 10 pmol of each primer, 1.5 U of Taq polymerase and 3 μl of template DNA. PCR amplification was carried out using thermal cycler (Techne-TC-512) with the following thermal regime; initial denaturing for 5 min at 94°C, 34 cycles of denaturing at 94°C for 40 s (BOX) and 50 s (ERIC), followed by annealing at 50°C for 40 s and 52°C for 90 s for BOX and ERIC primers respectively, elongation at 72°C for 60 s (BOX) and 240 s (ERIC) and final extension step at 72°C for 10 min. After finishing the PCR reactions, 7 μl of amplified products were mixed with 2 μl of loading buffer (Dye) and were electrophoresed through a 1.5% agarose gel in 1x TBE buffer at 80 V for 90 min and stained with ethidium bromide (0.5 μg/ml), and visualized under UV light using gel documentation. The Gene Ruler 1 kb DNA Ladder (Ready to Load, Solis BioDyne) was used to determine fragment size.
Data analysis
Clustering of bacterial isolates in this study was performed with comparison of phenotypic and biochemical characteristics and also based on fingerprints of genomic DNA of Pcc isolates in agarose gel. The rep-PCR banding patterns of agarose gel were scored as present (1) or absent (0) for each rep-PCR. Also, each physiological or biochemical characteristic was counted as a unit character; positive (1) or negative (0) test results were scored as binary traits. Cluster analysis and dendrogarm drawing were done by using the NTSYS software (version 2.02; Exeter Software, USA), using Jaccard coefficient and simple matching according to the unweighted pair group method with arithmetic averages (UPGMA) (Sutra et al., 2001). For phylogenetic analysis, 16S rRNA gene sequences were aligned with Geneious software (Java Version 1.7.0_51-b13) and the phylogenetic tree was created using Geneious Tree Builder software according to genetic distance model Tamura Nei, Neighbour-joining (NJ) method, and bootstrap 500.
Nucleotide sequence accession numbers
The 16S rDNA sequences from Pectobacterium CbUr23, CbUr22, PtUr26 and BpUr43 strains in this study were deposited in the GenBank database under the following accession numbers: KM371724, KM371725, KM371726 and KM371727 respectively.
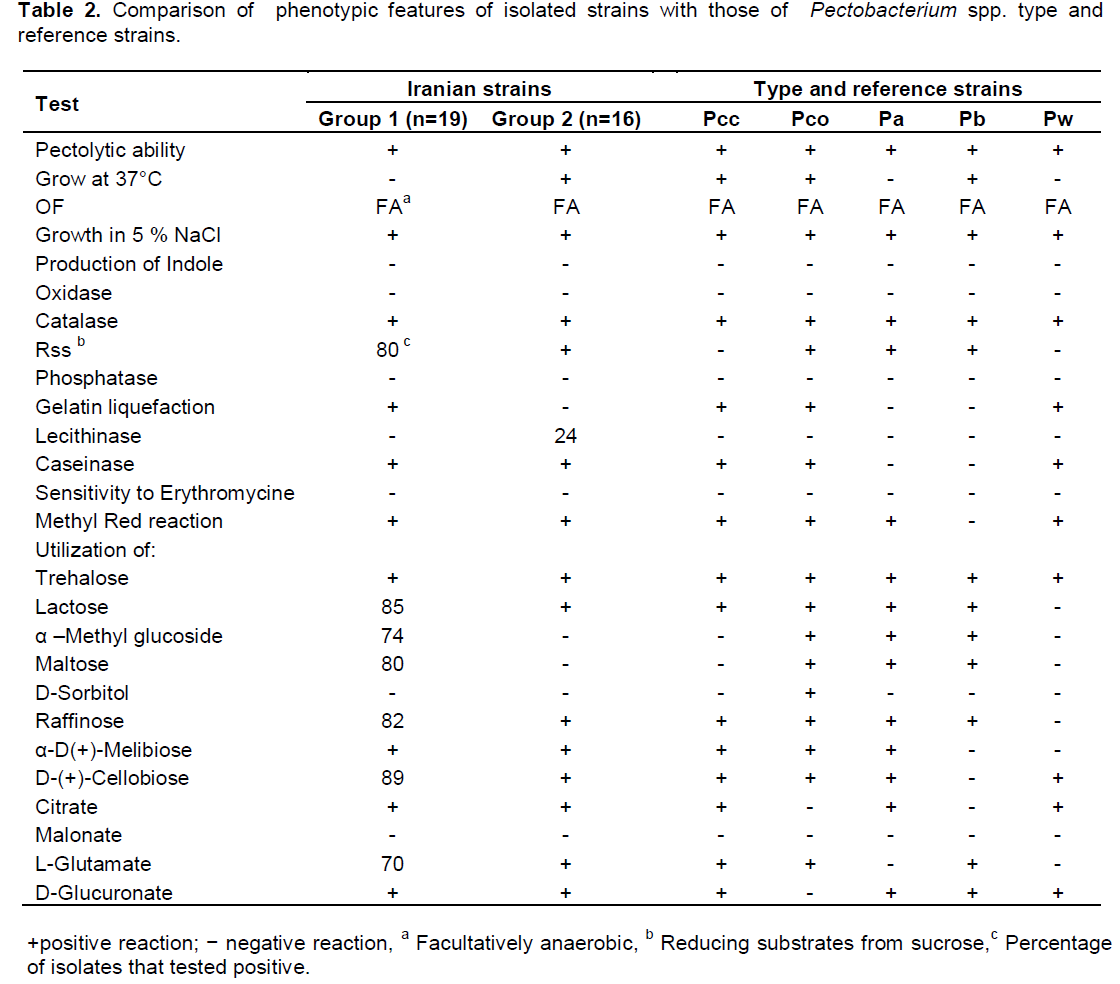
Phenotypic and biochemical characteristics
A survey was performed on infected vegetable tissues such as potato tuber, carrot root, bell pepper fruit and cabbage leaf in 2012-2013. In total, 35 isolates from four vegetable crops from north-west of Iran were all identified as Pectobacterium spp. by biochemical and phenotypic assays. The physiological and biochemical tests divided the isolates into two main groups according to their ability to grow at 37°C (Table 2). In addition, these two groups were differentiated by utilization of α –methyl glucoside and maltose, reducing substrates from sucrose and gelatin liquefaction. All bacterial cultures isolated from the survey exhibited pectolytic ability on potato slices. The isolates were also negative for indole production, phosphatase activity, acid production from maltose, utilization of malonate and urease activity. All the isolates were positive caseinase activity, methyl red reaction and utilization of trehalose, α-D-melibiose, citrate, L-glutamate and D-glucuronate. The biochemical tests for production of phosphatase and indole, sensitivity to erythromycin, growth in sodium chloride 5% agreed with those expected for Pectobacterium spp. (Gallois et al., 1992; Gardan et al., 2003). Therefore, based on biochemical features, all of the isolates of group 2 were determined as P. carotovoum subsp. carotovorum. Whereas, the isolates of group 1 showed heterogenous biochemical and physiological characteristics. All of the six isolates obtained from bell pepper belonged to group 1 were identified as P. wasabiae and they could be differentiated by other biochemical tests such as reducing substances from sucrose, utilization of lactose, and raffinose that were negative for P. wasabiae. Only four isolates from infected potatoes belonged to group 1 were identified as P. atrosepticum. The rest of group 1 initially described as atypical P. carotovoum subsp. carotovorum because they did not grow at 37°C. Despite this, there were five Pectobacterium spp. strains that their phenotypic and biochemical characteristics were not fitted well with the results obtained from type strains.
Pathogenicity test and hypersensitive response
The pathogenicity of representative strains of potato, bell pepper, carrot and cabbage was quantified using tuber, fruit, root and leaf of these vegetable crops respectively. All inoculated potato tubers, bell pepper fruits, carrot roots and cabbage leaves exhibited soft rot symptoms after 72 h similar to those observed in the fields and stores and the same bacteria were consistently re-isolated. Symptoms were not observed on water-inoculated controls. In the pathogenicity assay of potato tubers and carrot roots, both groups 1 and 2 produced small water soaked lesions or soft rot symptoms by the end of 48 h after inoculation. Disease symptoms on bell pepper fruits initiated by water-soaked lesions, and crumpled skin near the inoculation site after 48 h that extended gradually. At the same period, water-soaked and dark-green lesions appeared on cabbage leaves that were rotten rapidly. Strains of both groups showed the same symptoms and severity in potato maceration and pathogenicity on host plants assays. No signs of HR reaction or tissue collapse were observed in areas injected with any of the Iranian Pectobacterium species on tobacco 24 h after inoculation. As expected, HR reaction signs were not visible on tobacco injected with distilled water as negative control.
Analysis of 16S rRNA gene of the isolates
To confirm the results obtained by physiological and biochemical assays, we analyzed the partial 16S rRNA gene sequence by PCR amplification using the primer sets described in materials and methods. The PCR amplified product had approximately the size of 1200 bp when electrophoresed on 1% agarose gel (Figure 1). When the PCR products obtained using specific primers were sequenced and analyzed by comparison with available sequences in the GenBank database, BpUr43 16S rRNA gene was 99% homologous to the P. wasabiae strain WPP163.
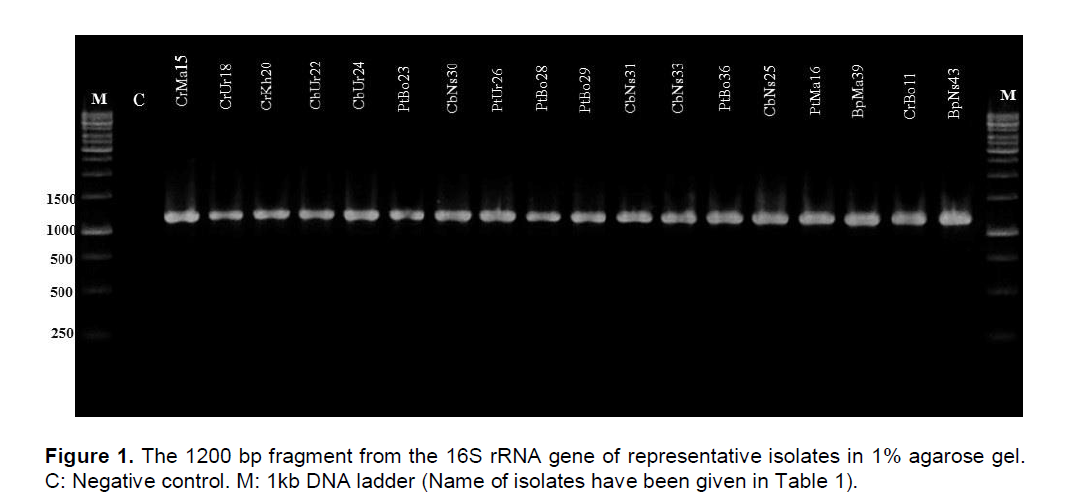
The PtBo23 16S rRNA gene exhibited 99% homology to the 16S rRNA gene of P. atrosepticum strain CFBP 1526. Whereas the strains CbUr22 and PtUr26 16S rRNA gene exhibited 99% homology to the 16S rRNA gene of P. carotovorum strain ATCC 15713 and P. carotovorum subsp. carotovorum strain ATCC 15713 respectively. The phylogenetic tree of the isolates obtained by Neighbour-joining (NJ) and Maximum Likelihood (ML) methods using MEGA 6 software (Figure 2). Pseudomonas aeruginosa used as outgroup in this study. Partial sequence of PCR product from reaction of putative Pectobacterium spp. with 16S rRNA confirmed the results obtained from physiological and biochemical assays used for identification of the bacterium. Application of specific primers such as PCCSSF/PCCSSR successfully differentiated Iranian P. wasabiae and P. carotovorum subsp carotovorum isolates from other species and subspecies of Pectobacterium.
Rep-PCR genomic fingerprinting of isolated Pcc strains
DNA amplification of Pcc strains and standard isolate were performed by rep-PCR method using two BOX and ERIC primers and they were assessed for their abilities to differentiate between isolated strains of P. carotovorum subsp. carotovorum. The rep-PCR banding patterns of each gel were normalized by Alpha Ease Fc 4.0 (Alpha Innotech of San Leandro, Calif.). Normalized bands were scored as present (1) or absent (0) for each rep-PCR. Individual and combined finger prints of two rep-PCR were analyzed by Pearson correlation and UPGMA using NTSYSpc software (ver 2.0; Exeter Software, USA). On this basis, with the use of BOX-PCR amplified DNA, fingerprints ranged from nine to 18 bands with sizes between 200 and 2,500 bp. While ERIC-PCR produced 11 to 20 bands with sizes between 180 and 4,500 bp (Figure 3).
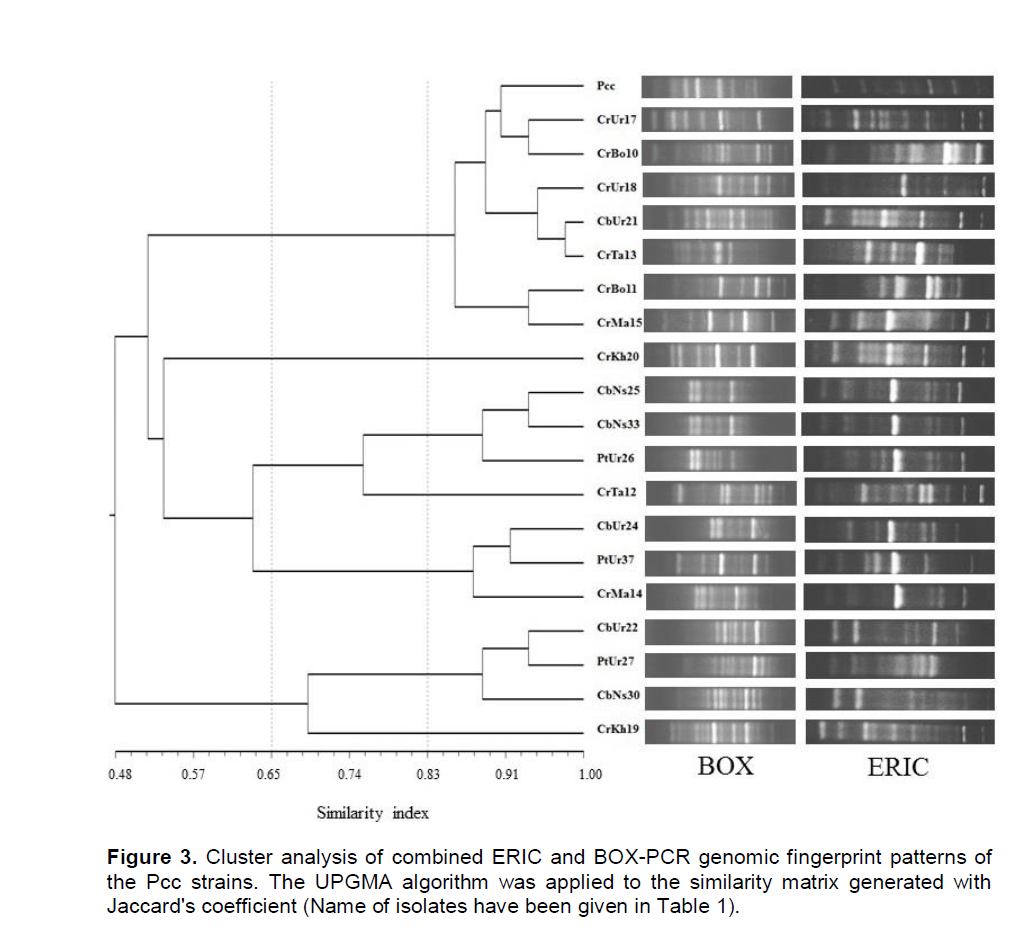
Based on UPGMA, BOX-PCR and ERIC-PCR individual profiles revealed three main clusters at 58 and 55.5% similarity value respectively and were subdivided the Pcc strains to five clusters with a similarity coefficient of 65%. Whilst cluster analysis of combined finger-print patterns of two rep-PCR revealed three clusters with a similarity coefficient of 53%, which were subdivided to five and seven clusters with a similarity coefficient of 65 and 83% respectively (Figure 3).
Our research goal was to identify Pectobacterium spp. as the causal organisms of soft rot through phenotypic and molecular techniques. A polyphasic approach including physiological, biochemical and molecular characteristics was used in the present investigation to identify isolated strains of soft rot causing Pectobacterium species from carrot, potato, bell pepper, and cabbage in north-west of Iran.
We examined two methods to differentiate the Pectobacterium isolates and found that phylogenetic analysis of the 16S rRNA gene was the most accurate method to characterize our set of pectolytic enterobacteria. Our results have demonstrated that all of the isolated bacteria from infected tissues (35 isolates) belonged to Pectobacterium spp. Most of the isolates were identified as P. carotovorum subsp. carotovorum using biochemical and physiological experiments. The existence of some unusual Pcc strains from potato, carrot and cabbage (group 1) that were unable to grow at 37°C and their inability to elicit HR on tobacco leaves persuaded us to check these isolates with analysis of the 16S rRNA gene. We applied PCR-based technique with specific primer for Pectobacterium sp. to enhance detection simplicity and rapidity in comparison to physiological and biochemical assays. However, all the strains were able to amplify specific bands relevant to Pectobacterium spp. Diversity in phenotypic characteristic among isolated Pcc strains from different plant hosts within the same geographical regions have been commonly reported in Golkhandan et al. (2013); Baghaee et al. (2011); Toth et al. (2011); Hu et al. (2008); Duarte et al. (2004) and Gardan et al. (2003).
Pectolytic Erwinias do not have specific host range and they can cause soft rot disease in broad host range and vast geographical distribution (Barras et al., 1984). The pathogenicity of obtained isolates from one host on another plants confirmed this subject. The virulence of representative strains of carrot and bell pepper were higher than other isolates.
P. carotovorum subsp carotovorum was previously reported as the major soft rotting causal agent on vegetable crops and ornamental plants in Iran (Soltani-Nejad et al., 2005; Firoz et al., 2007; Baghaee et al., 2011). Recently in parts of Iran another species of soft rotting bacteria was observed which was different from Pcc in some aspects (Baghaee et al., 2011). These isolates, unlike typical Pcc strains could not grow at 37°C and elicit HR on tobacco leaves.
In agreement with Yap et al. (2004); Kim et al. (2009); Pitman et al. (2009) and Baghaee et al. (2011), the members of group 1 including atypical Pcc strains from potato were confirmed as P. wasabiae by using a combination of physiological, biochemical and phylogenetic analysis. This is the first time that the presence of P. wasabiae strains from bell pepper in Iran has been described. Baghaee et al. (2011) isolated P. wasabiae strains from potato which elicited HR on tobacco, while our findings were in agreement with many other studies by Moleleki et al. (2012) and Glasner et al. (2008) that showed P. wasabiae was not able to elicit HR on tobacco. Even though in this aspect putative P. wasabiae strains were similar to reference strains of P. atrosepticum (SCRI 1043), and some atypical Pcc, they were differentiated from these two species of Pectobacterium by utilization of raffinose and lactose. Similar to other researches, P. wasabiae isolates of this study were able to cause soft rot of tubers as well as stem lesions (Yap et al., 2004; Pitman et al., 2009; Baghaee et al., 2011). Therefore, it could be hypothesized that different pectolytic species may use different mechanisms to overcome plant barriers.
Genetic diversity of Pcc strains in this study was also performed based on rep-PCR. Comparing the results of genetic fingerprint BOX-PCR and ERIC-PCR, some similarities and differences were observed in their classification. In both cases, strains were placed in three main fingerprinting groups. There was no complete correspondence between the BOX-PCR results and ERIC-PCR and even those strains which showed the same genotype in BOXPCR showed different genotypes in ERIC-PCR. Geographically, the groups created by BOX-PCR were closely related to their sample collection areas. In ERIC-PCR, these correlations were defined. Linkage between rep-PCR results and the geographic origin of bacterial strains has been recognized in various studies (Scortichini et al., 2001; Mkandawire et al., 2004). Louws et al. (1994) believe that one of the important reasons for this phenomenon is that the selection for one geographically suitable area can have influence on the genetic map of bacterium and also dispersion of these repetitive units in the genome of bacterium. This work supports this idea and in some cases artifacts, these relations were observed. No doubt rep-PCR is a reliable tool for epidemiological studies of diseases and one can use the information as a device for detection of pathogens. Despite this, more recent work with ISSR-PCR of bacteria such as Clavibacter michiganensis subsp. michiganesisis proved the greater sensitivity, specificity and reliability of this technique as another helpful informative tool in epidemiological studies (Baysal et al., 2011).
Moreover, in this study, we have shown that a polyphasic approach including physiological, biochemical and molecular characteristics is relatively a reliable method to classify strains of Pectobacterium spp. However, it seems that phylogenetic analysis with housekeeping genes can be the most accurate method to characterize strains of pectolytic enterobacteria. In summary, we can conclude that the damaging enterobacteria isolates from bell pepper storages in north-west of Iran belong to P. wasabiea. While, in this area the most of damaging enterobacteria strains isolated from carrot, potato and cabbage belong to P. carotovorum subsp carotovorum.
The authors have not declared any conflict of interests
REFERENCES
|
Agrios GN (2005). Plant Pathology. Department of Plant Pathology. University of Florida 948p.
Crossref
|
|
|
|
Alippi AM, Lopez AC (2009). First Report of Pectobacterium carotovorum subsp. carotovorum on Spathiphyllum wallisii in Argentina. Plant Dis. 93:842.
Crossref
|
|
|
|
|
Ayers SH, Rupp P, Johnson WT (1919). A study of the alkali-forming
Crossref
|
|
|
|
|
Baghaee-Ravari S, Shams-Bakhsh M, Rahimian H, Lopez-Solanilla E, Antúnez-Lamas M, Rodríguez-Palenzuela P (2011). Characterization of Pectobacterium species from Iran using biochemical and molecular methods. Eur. J. Plant Pathol. 129:413-425.
Crossref
|
|
|
|
|
Barras F, Van Gijsegem F, Chatterjee AK (1984). Extracellular enzymes and pathogenesis of soft-rot Erwinia. Ann. Rev. Phytopathol. 32:201-234.
Crossref
|
|
|
|
|
Bauer DW, Bogdanove AJ, Beer SV, Collmer A (1994). Erwinia chrysanthemi hrp genes and their involvement in soft rot pathogenesis and elicitation of the hypersensitive response. Mol. Plant Microbe Interact. 7:573-581.
Crossref
|
|
|
|
|
Baysal Ö, Mercati F, Ikten H, Yıldız RÇ, Carimi F, Aysan Y, Teixeira da Silva JA (2011). Clavibacter michiganensis subsp. michiganesis: Tracking strains using their genetic differentiations by ISSR markers in Southern Turkey. Physiol. Mol. Plant Pathol. 75:113-119.
Crossref
|
|
|
|
|
Boyraz N, Batas, KK, Maden S, Yasar A (2006). Bacterial Leaf and Peduncle Soft Rot Caused by Pectobacterium carotovorum on Tulips in Konya, Turkey. Phytoparasitica 34:272-280.
Crossref
|
|
|
|
|
Chao YC, Feng CT, Ho WC (2006). First report of Aglaonema bacterial blight caused by Erwinia chrysanthemi in Taiwan. Plant Dis. 90:1358.
Crossref
|
|
|
|
|
Cheng HR, Jiang N (2006). Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol. Lett. 28:55-59.
Crossref
|
|
|
|
|
Dickey RS, Kelman A (1988). Erwinia 'Carotovora' or soft rot group. In N. W. Schaad (Ed.), Laboratory guide for identifcation of plant pathogenic bacteria, 2nd edn, St Paul, MN: American Phytopathological Society pp. 44-46.
|
|
|
|
|
Duarte V, De Boer SH, Ward LL, de Oliveria AMR (2004). Characterization of a typical Erwinia carotovora strains causing blackleg of potato in Brazil. J. Appl. Microbiol. 96:535-545.
Crossref
|
|
|
|
|
Fahy PC, Persley GJ (1983). Plant Bacterial Disease: A Diagnostic Guide. Academic Press, Sydney, Australia.
|
|
|
|
|
Farrar JJ, Nunez JJ, Davis RM (2000). Influence of soil saturation and temperature on Erwinia chrysanthemi soft rot of carrot. Plant Dis. 84:665-668.
Crossref
|
|
|
|
|
Firoz R, Bahar M, Sarif-Nabi B (2007). Detection of casual agents of potato soft rot and blackleg in Esfehan province. Iran. J. Plant Pathol. 43:145-162.
|
|
|
|
|
Gallois A, Samson R, Ageron E, Grimont PA (1992). Erwinia carotovora subsp. odorifera subsp. nov. associated with odorous soft rot of chicory. Int. J. Syst. Bacteriol. 42:582-588.
Crossref
|
|
|
|
|
Gardan L, Gouy C, Christen R, Samson, R (2003). Elevation of three subspecies of Pectobacterium carotovorum to species level: Pectobacterium atrosepticum sp. nov., Pectobacterium betavasculorum sp. nov. and Pectobacterium wasabiae sp. nov. Intl. J. Syst. Evol. Microbiol. 53:381-391.
Crossref
|
|
|
|
|
Gillings M, Holley M (1997). Repetitive element PCR fingerprinting (rep-PCR) using enterobacterial repetitive intergenic consensus (ERIC) primers is not necessarily directed at ERIC elements. Lett. Appl. Microbiol. 25:17-21.
Crossref
|
|
|
|
|
Glasner JD, Marquez-Villavicencio M, Kim HS, Jahn CE, Ma B, Biehl BS (2008). Niche-specificity and the variable fraction of the Pectobacterium pangenome. Mol. Plant Microbe Interact. 21:1549-1560.
Crossref
|
|
|
|
|
Golkhandan E, Kamaruzaman S, Sariah M, Zainal Abidin MA, Nasehi A (2013). Characterization of Malaysian Pectobacterium spp. from vegetables using biochemical, molecular and phylogenetic methods. Eur. J. Plant Pathol. 137:431-443.
Crossref
|
|
|
|
|
Goto M, Matsumoto K (1987). Erwinia carotovora subsp.wasabiae subsp. nov. isolated from diseased rhizomes and fibrous roots of Japanese horseradish. International J. Syst. Bacteriol. 37:130-135.
Crossref
|
|
|
|
|
Hu XF, Ying FX, Gao YY, Chen HM, Chen JS (2008). Characterization of Pectobacterium carotovorum subsp. carotovorum causing soft-rot disease on Pinellia ternata in China. Eur. J. Plant Pathol. 120:305-310.
Crossref
|
|
|
|
|
Hugh R, Leifson E (1953). The taxonomic significance of fermentative versus oxidative metabolism of carbohydrates by various Gram-negative bacteria. J. Bacteriol. 66:24-26.
|
|
|
|
|
Kim HS, Ma B, Perna NT, Charkowski AO (2009). Prevalence and virulence of natural type III secretion system deficient Pectobacterium strains. App. Environ. Microbiol. 75:4539-4549.
Crossref
|
|
|
|
|
Kim JH, Joen YH, Kim SG, Kim YH (2007). First report on bacterial soft rot of graft cactus Chamaecereus silvestrii caused by Pectobacterium carotovorum subsp. carotovorum in Korea. Plant Pathol. 23:314-317.
Crossref
|
|
|
|
|
Klement Z, Rudolph K, Sand DC (1990). Methods in phytobacteriology. Akademiai Kiado Budapest 540p.
|
|
|
|
|
Loreti S, Gallelli A, Zaccardelli M, Parisi M (2001). Characterization of isolates of Erwinia carotovora from artichoke. J. Plant Pathol. 83:236.
|
|
|
|
|
Louws FJ, Fulbright DW, Stephens CT, de Bruijn FJ (1994). Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl. Environ. Microbiol. 60:2286-2295.
|
|
|
|
|
Ma B, Hibbing ME, Kim HS, Reedy RM, Yedidia I, Breuer J, Glasner JD, Perna NT, Kelman A, Charkowsky AO (2007). Host range and molecular phylogenies of the soft rot entrobacterial genera Pectobacterium and Dickeya. Phytopathology 97:1150-1163.
Crossref
|
|
|
|
|
Maisuria VB, Nerurkar AS (2013). Characterization and differentiation of soft rot causing Pectobacterium carotovorum of Indian origin. Eur. J. Plant Pathol. 136:87-102.
Crossref
|
|
|
|
|
MacFadden LA (1961). Bacterial stem and leaf rot of Dieffenbachia in Florida. Phytopathology 51:663-668.
|
|
|
|
|
Mkandawire AC, Mabagala RB, Guzman P, Gepts P, Gilbertson RL (2004). Genetic diversity and pathogenic variation of common blight bacteria (Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans) suggests pathogen coevolution with the common bean. Phytopathology 94:593-603.
Crossref
|
|
|
|
|
Moleleki LN, Onkendi EM, Mongae A, Kubheka GC (2012). Characterization of Pectobacterium wasabiae causing blackleg and soft rot disease in South Africa. Eur. J. Plant Pathol. 135(2):279-288.
Crossref
|
|
|
|
|
Nieves-Brum C (1985). Infection of roots of Dieffenbachia maculata by the foliar blight and soft rot pathogen, Erwinia chrysanthemi. Plant Pathol. 34:139-145.
Crossref
|
|
|
|
|
Perombelon MCM (1990). The genus Erwinia. In: The prokaryotes (Balows, A. Truper, H.G. Dworkin, M. Harder, W. and Schleifer, K. H. eds), 2nd edn, New York: Springer-Verlog. 3:2899-2921.
|
|
|
|
|
Perombelon MCM, Kelman A (1980). Ecology of the soft rot Erwinias. Ann. Rev. Phytopathol. 18:361-387.
Crossref
|
|
|
|
|
Perombelon MCM, Van der Wolf JM (2002). Methods for the detection and quantification of Erwinia carotovora subsp. atroseptica (Pectobacterium carotovorum subsp. atrosepticum) on potatoes: A laboratory manual. Dundee, Scotland: Scottish Crop Research Institute Occasional Publication No. 10.
|
|
|
|
|
Pitman AR, Harrow SA, Visnovsky SB (2009). Genetic characterisation of Pectobacterium wasabiae causing soft rot disease of potato in New Zealand. Eur. J. Plant Pathol. 126:423-435.
Crossref
|
|
|
|
|
Samson R, Lehendre JB, Achouak W, Gardan L (2005). Transfer of Pectobacterium chrysanthemi (Burkholder et al., 1953) Brenner et al., 1973 and Brenneria paradisiacal to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and delineation of four novel species, Dickeya dadantii sp. nov., Dickeya diffenbachiae sp. nov. and Dickeya zeae sp. nov. Int. J. Syst. Evol. Microbiol. 55:1415-1427.
Crossref
|
|
|
|
|
Schaad NW, Jones JB, Chun W (2001). Laboratory Guide for Identification of Plant Pathogenic Bacteria. Am. Phytopathol. Soc. St. Paul Minnesota 373p.
|
|
|
|
|
Scortichini M, Marchesi U, Di Prospero P (2001). Genetic diversity of Xanthomonas arboricola pv. juglandis (synonyms: X. campestris pv. juglandis; X. juglandis pv. juglandis) strains from different geographical areas shown by repetitive polymerase chain reaction genomic fingerprinting. J. Phytopathol. 149:325-332.
Crossref
|
|
|
|
|
Soltani-Nejad S, Taghavi M, Hayati J, Mostofi Z-G R (2005). Study of phenotypic and pathogenicity characteristics of Pectobacterium causing soft rot in Khozestan province. Iran. J. Plant Pathol. 41:585-611.
|
|
|
|
|
Sutra L, Christen R, Bollet C, Simoneau P, Gardan L (2001). Samsonia erythrinae gen. nov., sp. nov., isolated from bark necrotic lesions of Erythrina sp., and discrimination of plant-pathogenic Enterobacteriaceae by phenotypic features. Int. J. Syst. Evol. Microbiol. 51:1291-1304.
Crossref
|
|
|
|
|
Toth IK, Avrova AO, Hyman LJ (2001). Rapid identification and differentiation of the soft rot erwinias by 16S-23S intergenic transcribed spacer-PCR and restriction fragment length polymorphism analyses. Appl. Environ. Microbiol. 67:4070-4076.
Crossref
|
|
|
|
|
Toth IK, van derWolf JM, Saddler G, Lojkowska E, Hèlias V, Pirhonen M, Tsror Lahkim L, Elphinstone JG (2011). Dickeya species: an emerging problem for potato production in Europe. Plant Pathol. 60:385-399.
Crossref
|
|
|
|
|
Van der Merwe JJ, Coutinho TA, Korsten L, Van der Waals E (2010). Pectobacterium carotovorum subsp. Brasiliensis causing blackleg of potatoes in South Africa. Eur. J. Plant Pathol. 126:175-185.
Crossref
|
|
|
|
|
Versalovic J, Koueth T, Lupski JR (1991). Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acid Res. 19:6823-6831.
Crossref
|
|
|
|
|
Weingart H, Volksch B (1997). Ethylene production by Pseudomonas syringae pathovars in vitro and in planta. Appl. Environ. Microbiol. 63:156-161.
|
|
|
|
|
Yahiaoui-Zaidi R, Jouan B, Andrivon D (2003). Biochemical and molecular diversity among Erwinia isolates from potato in Algeria. Plant Pathol. 52:28-40.
Crossref
|
|
|
|
|
Yanagi M, Yamasato K (1993). Phylogenetic analysis of the family Rhizobiaceae and related bacteria by sequencing of 16SrRNA gene using DNA sequencer. FEMS Microbiol. Lett. 107:115-120.
Crossref
|
|
|
|
|
Yap MN, Barak JD, Charkowski, AO (2004). Genomic diversity of Erwinia carotovora subsp. carotovora and its correlation with virulence. Appl. Environ. Microbiol. 70:3013-3023.
Crossref
|
|
|
|
|
Zohour Paralak E, Rahimian H, Banihashemi Z (2007). A comparative study on pectolytic erwinias isolated from potato in the Fars province. Iranian J. Plant Pathol. 43:121-144.
|
|