ABSTRACT
Information about genetic variation has become more critical and deciding factor for any breeding and improvement effort,is difficult inefficient and inaccurate when based on morphological traits only. Therefore, this investigation was conducted using nine microsatellite DNA markers to evaluate genetic variation among 96 muskmelon germplasm in Bangladesh. All nine microsatellite markers were polymorphic. Lower values detected in observed than expected heterozygosity for all loci, indicating homozygous condition in the sample population. A total number of 28 alleles were generated by the nine primers across 96 germplasm. The locus TJ10 showed the highest number of alleles (5), whereas CMGA104, CMCT44, CMAG59, CMTA134a and J27 were generated lowest (3) number of alleles.Allele size ranged between 98 bp (CMCT44) to 198 (CMCTT144). The Polymorphism Information Content (PIC) valuesranged from 0.117 to 0.770for the locus CMCTN86and TJ10, repectively. Values of genetic differentiation (Fst) and gene flow (Nm) ranged from 0.535 to 1.000 with an average of 0.776 and from 0.000 to 0.218 with a mean value of 0.072, respectively. Broad genetic base was found among the muskmelon germplasm used in this study. The average pair-wise Nei’s genetic distance value was 0.605. The highest genetic dissimilarity coefficient (GD=2.300) was between the germplasm AHM-241 and IAH-102, whereas germplasm IAH-251 and IAH-259 comprised lowest genetic diversity (GD =0.000). In the UPGMA dendrogram, among 96 germplasm of muskmelon 84 grouped in cluster “I” and other 12 in cluster “II”.
Keywords: Muskmelon, Microsatellite (SSR) marker, Polymorphism, Genetic diversity, Germplasm
The muskmelon (Cucumis melo L., 2n=24) is one of the most nutritive and commercially important cucurbit in the world. Generally, it is considered as a crop of tropical and subtropical regions, extensive cultivation observed in temperate climates. Desert and savannah regions of Africa, Arabia, southwestern Asia and Australia are the origin of wild populations of muskmelons. Based on the muskmelon largest consumers across the world, the United States is one of them. According to Boriss et al. (2014), Americans consume 27 pounds per year of melons in an average; in which 8.7 pounds is cantaloupe. Muskmelon is normally eaten as a fresh fruit, salad, or dessert with ice cream or custard. In Bangladesh, people consume both unripe and ripe fruits. Unripe fruits are consumed as salad and processed food as soup, stew, curry, stir-fry or pickle. Mature ripe fruits are eaten fresh as a desert fruit and sometimes slightly processed as canned, syrup, jam or dehydrated slices (Malek et al., 2012). Moreover, it is a source of polyphenol antioxidants, chemicals which can regulate the formation of nitric oxide, a vital chemical for prevention of heart attacks.
In Bangladesh, it is considered as a minor but the most common fruit crop of Cucurbitaceae family. It is cultivated all over the country in Bangladesh. According to the latest statistics provided by BBS (2016), it was indicated that the area and production of muskmelon in Bangladesh are 4047 ha and 53000 tons, respectively and contributes 1.13% to total fruit production in Bangladesh.
Despite the variation in habit, size, shape, colour, maturity time and yield observed in Bangladeshi muskmelon germplasm, very little work has been done on genetical improvement of this crop. Muskmelon being predominantly andromonoecious, is a cross pollinated crop and provide ample scope for utilization of the hybrid vigor. Assessment of genetic variability and selection of suitable genotypes is the foremost criterion for genetical improvement of crop species. Plant Genetic Resources Centre (PGRC) of Bangladesh Agricultural Research Institute (BARI) has conserved different types of muskmelon germplasm collected from different parts of Bangladesh. Islam et al. (2017) reported that a total of 131 germplasm of muskmelon were collected and conserved within 2015 to 2016 in PGRC, BARI. Among these collected germplasm, diverse germplasm could be used as a source of genetic material for improving the yield, earliness, uniformity, quality and resistance to biotic and abiotic stresses in muskmelon. Conventionally, morphological markers called descriptors were used for varietal identification and genetic diversity analysis in plants which is time-consuming and expensive, requiring large areas of land and skilled personnel, and are often subjective due to environmental influences (Singh et al., 2004). However, the level of polymorphism for morphological characteristics in elite germplasm is sometimes too limited and inadequate to allow for variety/genotype discrimination (Geleta et al., 2004). In that situation, DNA marker is a one stop solution to combat these problems.
Several researchers have used successfully a variety of DNA markers to characterize the genetic diversity of melons such as isozymes (Staub et al., 1997; Akashi et al., 2002), restriction fragment length polymorphism (RFLPs) (Zheng et al., 1999), random amplification of polymorphic DNAs (RAPDs) (Garcia et al., 1998; Stepansky et al., 1999; Mliki et al., 2001; Lo´pez-Sese et al., 2003; Staub et al., 2004; Sensoy et al., 2007; Tanaka et al., 2007, Nhi et al., 2010; Soltani et al., 2010), amplified fragment length polymorphism (AFLPs) (Garcia-Mas et al., 2000, Yashiro et al., 2005), inter-simple sequence repeat (ISSR) and simple-sequence repeat (SSR) (Katzir et al., 1996; Staub et al., 2000; Danin-Poleg et al., 2001; Lo´pez-Sese´ et al., 2002; Monforte et al., 2003; Nakata et al., 2005; Tzitzikas et al., 2009; Raghami et al., 2014; Trimech et al., 2015) using diverse germplasm from different locations worldwide. Of all classes of DNA based marker, SSR markers represented by the repeats of 1-6 nucleotide-long DNA motifs arranged in tandem, have been considered one of the most powerful Mendelian markers (Jarne and Lagoda, 1996) because of their high reproducibility, co-dominance inheritance, multi-allelic character, and extensive genome coverage (Powell et al., 1996). The polymorphism of SSRs, primarily resulting from the variation of repeat numbers, can be easily detected by a simple PCR technique. In this context, the aim of the present study is to determine the genetic diversity of the muskmelon germplasm in Bangladesh and to find out the phylogenetic relationships among the 96 germplasm using Simple Sequence Repeat (SSR) markers.
Plant materials
Ninety six germplasm collected from different sources were used as plant materials (Table 1). However, variation among these germplasm based on fruit skin colour, size and shape are given in Figure 1.
Plant sample and extraction of genomic DNA
Diversity at molecular level was studied at the Molecular Biology Laboratory, Plant Genetic Resources Centre of Bangladesh Agricultural Research Institute, Gazipur using SSR markers. Young, fresh, disease and insect free leaves were used for DNA extraction. The genomic DNA was isolated from a bulk of 3-week old seedling leaf tissues taken from 5 plants from each germplasm using SDS and phenol: chloroform: IAA followed by alcohol precipitation described by Saghai-Maroof et al. (1984) with some modifications. Apart from usage of liquid nitrogen, the leaf sample was cut into
small pieces and digested with homogenization buffer containing Tris-50 mM, EDTA-25 mM, NaCl-300 mM, 1% SDS and deionized water was used. It was incubated at 65ºC for 30 min, extracted with phenol: chloroform: isoamyl alcohol (25:24:1), precipitated with ice-cold and extra pure isopropyl alcohol and purified with absolute ethanol (Plus sodium acetate, 3M) and 70% ethanol chronologically. DNA sample of each muskmelon germplasm was dissolved in 50 μl of TE buffer in 1.5-ml Eppendorf tube. When the DNA pellet was totally dissolved in TE buffer, 4 μl RNaseA (10 mg/ml) with isolated DNA was added and incubated at 37°C for 30 min (Tilahun et al., 2013). Finally, DNA sample was stored at -20°C.
Quantification and optimization of DNA concentration
The presence of genomic DNA was confirmed on 1% agarose gel qualitatively. The gels were visualized under UV light and photographed using photo documentation system (UV Transilluminator, Uvitec, UK). All of the DNA samples were found to be in good quality in this study. The amount of genomic DNA was quantified using UV spectrophotometer (Thermo Fisher Scientific, USA) at 260 nm. The original DNA concentrations of each muskmelon germplasm were measured following the formula described by Sumon et al. (2014).
Selection of microsatellite/SSR primers
Nine SSR primer pairs described previously (Katzir et al., 1996; ; Danin-Poleg et al., 2001; Henane et al., 2015) were used in the present study for microsatellite analysis (Table 2). All 9 primers pairs showed better responsiveness with clear and expected amplified product sizes.
PCR standardization and amplification
Amplification reactions were performed in 10-μL volumes containing 5X Green GoTaq Reaction Buffer (Promega, USA), 15 mM MgCl2, 1.25 U Taq DNA polymerase (Thermo Fisher Scientific, USA), 0.4 mM each of the dNTPs (NEB, USA), 10 μM forward and reverse primers and 50 ng template DNA. The mixtures were prepared at 0°C and transferred to the thermal cycler. Amplification reactions of SSR loci were carried out in a Mastercycler nexus Gradient thermal cycler (Eppendorf, Germany), using a program consisting of an initial denaturation step of 3 min at 94°C followed by 35 cycles of 45 s at 94°C, 1 min at 48 to 55°C and 1 min at 72°C; the program ended with a 8 min elongation step at 72°C. PCR products were stored at 4°C prior to analysis.
Gel electrophoresis and visualization of PCR products
PCR-products were electrophoresed on a 5% denaturing polyacrylamide gel containing 19:1 acrylamide: Bis-acrylamide, 10X TBE buffer, 10% APS and UltraPure TEMED. Electrophoresis was done using the Triple Wide Mini-Vertical Electrophoresis System, MGV-202-33 (CBS Scientific, USA). The gel was run at 80 to 90 V for a specified period of time depending on the size of amplified DNA fragment (usually 1 h for 100 bp). Upon loading of PCR products, 20°C temperature was maintained by a cooling system (Julabo, Germany). When electrophoresis was completed, the gel was stained with ethidium bromide (10 mg/ml) for 30 min. Finally, the stained gel was soaked in deionized water for 5 min. The individual bands were visualized under UV light and scored for analysis (Sumon et al., 2014).
Preparation of microsatellite data matrix for analysis
SSR markers were attained due to codominant nature microsatellite markers; it could be applied to distinguish both homozygous and heterozygous genotypes in individual plants. Individual alleles (bands) at the microsatellite loci were scored and single data matrix constructed for all loci as described by Molla et al. (2016). The constructed data matrix was used to calculate statistics of genetic variation and cluster analyses were carried out based on the genetic distance (Nei, 1972); unweighted pair group method with arithmetic mean (UPGMA) dendrogram constructed using computer program POPGENE (Version 1.31) (Yeh et al., 1999) and the relationships among the germplasm was determined. The PIC value was calculated using PIC = 1- Σf2ij; where fij is the frequency of the ith allele for the jth SSR locus (Anderson et al., 1993). PIC values express the discriminating ability of the particular. These values varied based on the number of alleles per locus and the relative frequencies of those alleles in the population. Allelic lengths were estimated using the software DNA FRAG version 3.03 (Nash, 1991).
According to DNA amplification patterns, all nine microsatellite markers used in this analysis were found to be polymorphic. One typical SSR profile is shown in Figure 2. Analysis of the variability parameters for the 9 SSRs in the 96 muskmelon germplasm are shown in Table 3. A total of 28 alleles were identified among all muskmelon germplasm with the 9 SSRs loci scanned herein, with an average of 3.11 allele per locus, varying from two for ‘CMCTT144’ ‘CMTA170a’ and ‘CMCTN86’ to 5 for ‘TJ10’ loci (Table 3) which is lower than that reported by ; Danin-Poleg et al. (2001), and higher than the result obtained by Henane et al. (2015). Variation of allele sizes ranged from 98 to 198 bp. Highest number of observed alleles (5) were found at the locus TJ10 among the 96 muskmelon germplasm ranging in size from 141 to 160 bp followed by 4 alleles (132 to 157 bp and 98 to 113 bp) and 3 alleles (131 to 141 bp, 230 to 302 bp, 150 to 162 bp and 171 to 187 bp) at the loci CMGA104, CMCT44, CMAG59, CMTA134a, and J27, respectively. However, lowest number of alleles (2) size ranging from 184 to 198, 122 to 136 and 170 to 187 bp was detected for the loci ‘CMCTT144’ ‘CMTA170a’ and ‘CMCTN86’, respectively (Table 3). All the nine SSR loci were found to be polymorphic, proving their effectiveness for genetic analysis of muskmelon germplasm. Five alleles expected length of 124 bp for CMAG59 and 6 alleles expected length of 125 and 192 bp for CMGA104 and CMGA144, respectively in muskmelon cultivars were reported (Katzir et al., 1996; Danin-Poleg et al., 2001). Parallel observed allelic lengths were estimated compare to previous study although some variation raised might be due to mutation of di-nucleotide repeat units.
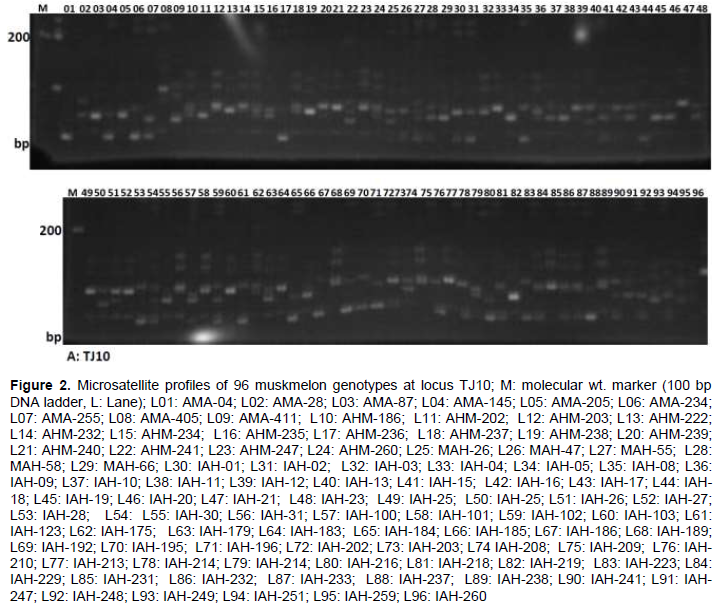
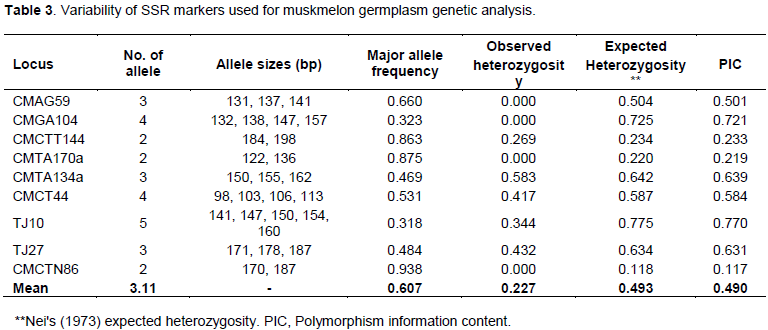
Heterozygosity can be considered as an indicator for the measurement of genetic variability. It expresses level of variation that exists in the population and how that variation is allocated across the alleles of an analyzed locus. Lower values of heterozygosity indicate small genetic variability. In general, small numbers of heterozygous individuals were observed for all SSRs, with an average of 0.227, ranging between 0.000 and 0.583 (Table 3). Expected heterozygosity (He, average 0.493) values for each SSR locus, considering all studied germplasm, were always higher than the observed heterozygosity (Ho), representing homozygous individuals in population samples. The observed heterozygosity is the proportion of heterozygous individuals in population samples; expected heterozygosity is the probability of an individual being heterozygous in any locus. In this study, the highest observed heterozygosity values (Ho=0.432) were attained with locus TJ27 (Table 3). Mean observed heterozygosity per locus in the studied germplasm tested herein was higher than those in Lo´pez-Sese´ et al. (2002) and Tzitzikas et al. (2009), probably due to the greater diverse germplasm used in the present study. Higher levels of heterozygosity would be expected because of primarily studied germplasm that had been developed by local farmers and, therefore cross-pollination might occur with other accessions. In this study, lower values of heterozygosity were observed, probably due to lack of intercrossing between them or with other accessions, a high rate of self-pollination. Another possibility is that the accessions originated from small populations or high levels of inbreeding (Raghami et al., 2014).
The PIC values represent the variation of allele and provide an estimation of discriminating ability of the marker. It is reflected by the number of alleles at a locus and also relative frequencies of these alleles. The genetic diversity of the studied germplasm might have an effect in variation of PIC values and high ratio of traditional cultivars used in this investigation might be a reason for raising the PIC values.
It is important to indicate that the selection by breeders have increased the frequency of the alleles or allelic combination with favorable effects at the expense of the others, eventually eliminating many of them (Cao et al., 1998). The estimated PIC values of nine SSR markers analyzed with 96 muskmelon germplasm were higher than zero. It is indicated that polymorphic and informative markers tested in this investigation was capable to describe genotypic variation of those germplasm. PIC values for nine SSRs ranged from 0.117 to 0.770 (Table 2), with an average of 0.490. Four of these SSRs were very informative (PIC > 0.6), with the highest PIC value recorded for TJ10 (0.770) and followed by CMGA104, CMTA134a and TJ27. Validations of informative markers depend on the level of polymorphism of this specific marker which is extremely useful for genetic studies (Sundaram et al., 2007).
Observed and effective number of alleles was also different in the present investigation. The mean number of observed allele and effective alleles for SSR loci with 96 muskmelon genotypes were 3.111 and 2.393, respectively (Table 4). These results were supported by previous study. For instance, 3.5 alleles using 30 SSR primers on 13 genotypes were reported by Danin-Poleg et al. (2001), while Lo´pez-Sese´ et al. (2002) found 2.4 alleles on 15 Spanish melons, and Tzitzikas et al. (2009) 2.47 alleles on 14 Greek and Cypriot melons. Monforte et al. (2003) detected 6.3 alleles on 27 wild and cultivated melons, which was divergent to the mean number of allele for reference genotypes in this study (3.111). This high value was due to various subspecies of melons which they examined.
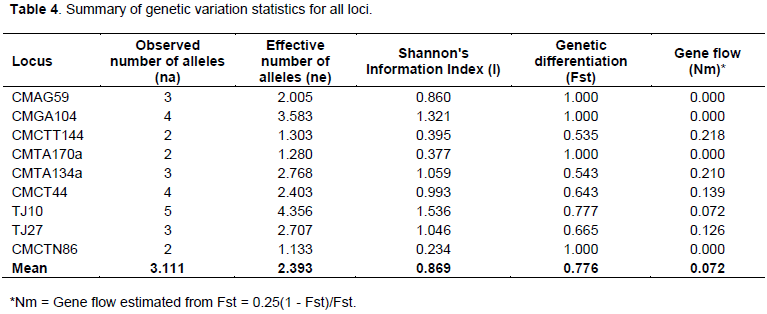
Genetic differentiation (Fst) values ranged from 0.535 to 1.000 with a mean value of 0.776 and gene flow (Nm) values were found in the ranges 0.000 to 0.218 with an average of 0.072 (Table 4). In this study, comparatively higher values of genetic differentiation and lower values of gene flow were observed among the nine SSR markers which are indicative of diversity among the genotypes as most of the studied genotypes were of land races and local cultivars. The mean Shannon’s information index (I) for all loci for 96 muskmelon germplasm were 0.869, and ranged from 0.046 to 1.000 (Table 4).
From these results, SSR markers can be used effectively to estimate genetic distances among genotypes. The mean genetic distance was 0.674 between Iranian cultivated melon (Raghami et al., 2014)and 0.285 between Spanish melon (Lopez-Sesé et al.,2002), while in the present study, it varied from 0.000 to 2.300 with an average of 0.605 (Figure 3). Different genetic background represents higher genetic distance values between germplasm pairs. However, least/nil values might be found in case of reverse genetic background. This variability in genetic distance values can play vital role for the enhancement of resources Bangladeshi muskmelon and sustainable use for genetical improvement. Genetic distance UPGMA dendrogram is built on SSR markers data referring to dissimilarity coefficient among the germplasm. On the basis of cluster study, the total genotypes were distributed into two main clusters; I and II. These two main clusters were further sub-divided into 17 sub-groups. Among these 17 sub-groups, 15 fell in main cluster I and the remaining two sub-groups were part of main cluster II (Figure 3). Upon subsequent separation, pair-wise estimates of dissimilarity ranged from 0.000 to 2.300 and the average similarity among all 96 germplasm was 0.605. Germplasm AHM-241 and IAH-102 had the highest genetic dissimilarity coefficient (GD=2.300) which was grouped in sub-cluster G6 and G17, respectively. Subgroup G6 gathered 15 genotypes in which genotypes IAH-251 and IAH-259 comprise sharp similarity (GD=0.000) that makes one think “synonymy phenomenon” may be the same germplasm, but having undergone two different names depending on the collection area. Similarly, diverse germplasm were grouped in different cluster in the dendrogram, because of variation in morphological traits and/or geographical distribution. For instance, germplasm AHM-241 and IAH-102 showed distinct variation in their fruit morphology (Figure 1).

Taken as a whole, the present study clearly show that studied germplasm with this broad genetic diversity could play an important role in the preservation and enhancement of muskmelon genetic diversity. Inter-mating genotypes from the major distinct gene pools could provide new genetic recombination to exploit in various development programme. In spite of the variability observed in this study, the 28 alleles of the 9 SSR loci were not adequate to discriminate all the 96 germplasm of muskmelon. For instance, AHM-222 vs. AHM-234 and IAH-175 vs. IAH-229 germplasm pair were genetically closely related for the loci analyzed. It is emphasized that these germplasm showed variation in some morphological traits like fruit skin, colour intensity, skin hardness of fruit, fruit shape and flesh colour (Islam et al., 2017). In situations where a set of pre-established markers were not able to differentiate accessions from a given species, Jakse et al. (2005) suggested that additional markers have to be used to reveal polymorphisms. Hence, it would be better to use higher number of primers for the creation and construction of an appropriate genetic relationship, germplasm identification and analysis of genetic variation. Moreover, it is necessary to use both morpho-physiological traits and molecular traits together to distinguish important germplasm for further genetical improvement of this crop species.
The authors have not declared any conflict of interests.
AFACI Secretariat, RDA, Republic of Korea, is greatly acknowledged for the financial support of this study through AFACI PAN-ASIAN PROJECT “Collection, Characterization and Promotion of Rice, Chilli, Cucumber and Melon in Bangladesh”
REFERENCES
|
Akashi Y, Fukuda N, Wako T, Masuda M, Kato K (2002). Genetic variation and phylogenetic relationships in East and South Asian melons, Cucumis melo L., based on the analysis of five isozymes. Euphytica 125:385-396.
Crossref
|
|
|
|
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrels ME (1993). Optimizing parental selection for genetic linkage maps. Genome 36:181-186.
Crossref
|
|
|
|
|
Bangladesh Bureau of Statistics (BBS) (2016). Yearbook of Agricultural Statistics-2015, Bangladesh Bureau of Statistics. Ministry of Planning, Government of the People's Republic of Bangladesh. P 42.
|
|
|
|
|
Boriss H, Brunke H, Keith M (2014). Melon Profile [Internet]. Agricultural Marketing Resource Center.
|
|
|
|
|
Cao T, Duprez E, Borden KLB, Freemont PS, Etkin LD (1998). Ret finger protein is a normal component of PML nuclear bodies and interacts directly with PML. J. Cell. Sci. 111:1319-1329.
|
|
|
|
|
Danin-Poleg Y, Reis N, Baudracco-Arnas S, Pitrat M, Staub JE, Oliver M, Aru’s P, de Vicente CM, Katzir N (2001). Development and characterization of microsatellite markers in Cucumis. Theor. Appl. Genet. 102:61-72.
Crossref
|
|
|
|
|
Garcia E, Jamilena M, Alvarez JI, Arnedo T, Oliver JL, Lozano R (1998). Genetic relationships among melon breeding lines revealed by RAPD markers and agronomic traits. Theor. Appl. Genet. 96:878-885.
Crossref
|
|
|
|
|
Garcia-Mas J, Oliver M, Gomez-Paniagua H, De Vicente MC (2000). Comparing AFLP, RAPD and RFLP markers for measuring genetic diversity in melon. Theor. Appl. Genet. 101:860-864.
Crossref
|
|
|
|
|
Geleta LF, Labuschagne MT, Vijoen CD (2004). Relationship between heterosis and Genetic distance based on morphological traits and AFLP markers in pepper. Plant Breed. 123:467-473.
Crossref
|
|
|
|
|
Henane I, Slimane RB, Jebari H (2015). SSR-based genetic diversity analysis of Tunisian varieties of melon (cucumis melo L.) and Fakous (cucumis melo var.flexuosus). Intl. J. Adv. Res. 3(3):727-734.
|
|
|
|
|
Islam MT, Malek MA, Islam MN (2017). Characterization of muskmelon germplasm (summer-2015-16). In: MT Islam, Afroz R, Rahman S and Molla MR (Editors), Annual Research Report on Plant Genetic Resources Conservation and Management 2016-17, Plant Genetic Resources Centre, Bangladesh Agricultural Research Institute, Joydebpur, Gazipur-1701. pp. 104-111.
|
|
|
|
|
Jakse J, Martin W, Mccallum J, Havey MJ (2005). Single nucleotide polymorphisms, indels, and simple sequence repeats for onion cultivar identification. J. Am. Soc. Hort. Sci. 130:912-917.
|
|
|
|
|
Jarne P, Lagoda PJL (1996). Microsatellites, from molecules to populations and back. Trends Ecol. Evol. 11:424-429.
Crossref
|
|
|
|
|
Katzir N, Danin-Poleg Y, Tzuri G, Karchi Z, Lavi U, Cregan PB (1996). Length polymorphism and homologies of microsatellites in several cucurbitacean species. Theor. Appl. Genet. 93:1282-1290.
Crossref
|
|
|
|
|
Lo’pez-Sese´ AI, JE Staub, ML Go’mez-Guillamo’n (2003). Genetic analysis of Spanish melon (Cucumis melo L.) germplasm using a standardized molecular marker array and reference accessions. Theor. Appl. Genet. 108:41-52.
Crossref
|
|
|
|
|
Lo’pez-Sese´ AI, Staub JE, Katzir N, Go’mez-Guillamo’n ML (2002). Estimation of between and within accession variation in selected Spanish melon germplasm using RAPD and SSR markers to assess strategies for large collection evaluation. Euphytica 127:41-51.
Crossref
|
|
|
|
|
Malek MA, Islam MO, Haque MM, Sultan MK (2012). Screening of muskmelon (Cucumis melo L.) germplasm against salinity. Bang. J. Agric. Res. 37(3):465-472.
Crossref
|
|
|
|
|
Mliki A, Staub JE, Sun ZY, Ghorbel A (2001). Genetic diversity in melon (Cucumis melo L.): an evaluation of African germplasm. Genet. Resour. Crop Evol. 48:587-597.
Crossref
|
|
|
|
|
Molla MR, Ahmed I, Rohman MM, Hossain MA, Chowdhury MAZ (2016). Genetic Diversity Analysis and DNA Fingerprinting of Mungbean (Vigna radiata L.) Genotypes Using SSR Markers. J. Plant Sci. 6(4):153-164.
|
|
|
|
|
Monforte AJ, Garcia-Mas J, Arus P (2003). Genetic variability in melon based on microsatellite variation. Plant Breed. 122:153-157.
Crossref
|
|
|
|
|
Nakata E, Staub JE, Lo’pez-Sese´ AI, Katzir N (2005). Genetic diversity of Japanese melon cultivars as assessed by random amplified polymorphic DNA and simple sequence repeat markers. Genet. Res. Crop. Evol. 52:405-419.
Crossref
|
|
|
|
|
Nash JHE (1991). DNAfrag, Version 3.03. Institute for biological sciences, National Research Council of Canada, Ottawa, Ontario, Canada.
|
|
|
|
|
Nei M (1972). Genetic distance between populations. Am. Nat. 106:283 292.
Crossref
|
|
|
|
|
Nei M (1973). Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 70:3321-3323.
Crossref
|
|
|
|
|
Nhi PTP, Akashi Y, Hang TTM, Tanaka K, Aierken Y, Yamamoto T, Chunlin LH, Kato K (2010). Genetic diversity in Vietnamese melon landraces revealed by the analyses of morphological traits and nuclear and cytoplasmic molecular markers. Breed. Sci. 60:255-266.
Crossref
|
|
|
|
|
Powell W, Machray GC, Provan J (1996). Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1:215-222.
Crossref
|
|
|
|
|
Raghami M, Lopez-Sese AI, Hasandokht MR, Zamani Z, Fattahi M, Mahmoud R, Kashi, A (2014). Genetic diversity among melon accessions from Iran and their relationships with melon germplasm of diverse origins using microsatellite markers. Plant Syst. Evol. 300:139-151.
Crossref
|
|
|
|
|
Saghai-Maroof MA, Biyashev RM, Yang GP, Zhang Q, Allard RW (1994). Extraordinarily polymorphic microsatellite DNA in barley: species diversity, chromosomal locations, and population dynamics. Proc. Natl. Acad. Sci. 91:5466-5470.
Crossref
|
|
|
|
|
Sensoy A, Büyükalaca S, Abak K (2007). Evaluation of genetic diversity in Turkish melons (Cucumis melo L.) based on phenotypic characters and RAPD markers. Genet. Resour. Crop. Evol. 54:1351-1365.
Crossref
|
|
|
|
|
Singh RK, Sharma RK, Singh AK, Singh VP, Singh NK, Tiwari SP, Mohapatra T (2004). Suitability of mapped sequence tagged microsatellite site markers for establishing distinctness, uniformity and stability in aromatic rice. Euphytica 136:135-143.
Crossref
|
|
|
|
|
Soltani F, Akashi Y, Kashi A, Zamani Z, Mostofi Y, Kato K (2010). Characterization of Iranian melon landraces of Cucumis melo L. groups Flexousus and Dudaim by analysis of morphological characters and random amplified polymorphic DNA. Breed. Sci. 60:34-45.
Crossref
|
|
|
|
|
Staub JE, Box J, Meglic V, Horejsi TF, McCreight JD (1997). Comparison of isozyme and random amplified polymorphic DNA data for determining intraspecific variation in Cucumis. Genet. Res. Cr. Evol. 44:257–269.
Crossref
|
|
|
|
|
Staub JE, Danin-Poleg Y, Fazio G, Horejsi T, Reis N, Katzir N (2000). Comparative analysis of cultivated melon groups (Cucumis melo L.) using random amplified polymorphic DNA and simple sequence repeat markers. Euphytica 115:225-241.
Crossref
|
|
|
|
|
Staub JE, López-Sesé AI, Fanourakis N (2004). Diversity among melon landraces (Cucumis melo L.) from Greece and their genetic relationship with other melon germplasm of diverse origins. Euphytica 136:151-166.
Crossref
|
|
|
|
|
Stepansky A, Kovalski I, Perl-Treves R (1999). Intraspecific classification of melons (Cucumis melo L.) in view of their phenotypic and molecular variation. Plant Syst. Evol. 217:313-332.
Crossref
|
|
|
|
|
Sumon MH, Habiba U, Bhuyan SI, Haque MS, Begum SN, Hossain MD (2014). DNA fingerprinting and genetic diversity analysis of chilli germplasm using microsatellite markers. Biotechnology 13:174-180.
Crossref
|
|
|
|
|
Sundaram GR, Venkatesan T, Kunnummal KV (2007). Genetic diversity among cultivars, landraces and wild relatives of rice as revealed by microsatellite markers. J. Appl.Genet, 48:337-345.
Crossref
|
|
|
|
|
Tanaka K, Akashi Y, Nishitani A, Sakata Y, Nishida H, Yoshino H, Kato K (2007). Molecular characterization of South and East Asian melon Cucumis melo L., and the origin of Group Conmon var. makuwa and var. conmon revealed by RAPD analysis. Euphytica 153:233-247.
Crossref
|
|
|
|
|
Tilahun S, Paramaguru P, Kannan Bapu JR (2013). Genetic diversity in certain genotypes of chilli and paprika as revealed by RAPD and SSR analysis. Asian J. Agric. Sci. 5(2):25-31.
|
|
|
|
|
Trimech R, Makram, AFIF, Boussaid, M (2015). Genetic diversity of Tunisian melon (Cucumis melo L) landraces and their relationships with introduced varieties as assessed by simple-sequence repeat (SSR) markers. Afr. J. Biotech. 14(2):86-95.
Crossref
|
|
|
|
|
Tzitzikas EN, Monforte AJ, Fatihi A, Kypriotakis Z, Iacovides TA, Ioannides IM, Kalaitzis P (2009). Genetic diversity and population structure of traditional Greek and Cypriot melon cultigens (Cucumis melo L.) based on simple sequence repeat variability. Hort. Sci. 44(7):1820-1824.
|
|
|
|
|
Yashiro K, Iwata H Akashi Y, Tomita K, Kuzuya M, Tsumura Y, Kato K (2005). Genetic relationship among East and South Asian melon (Cucumis melo L) revealed by AFLP analysis. Breed. Sci. 55:197-206.
Crossref
|
|
|
|
|
Yeh FC, Yang RC, Boyle T (1999). POPGENE VERSION 1.31: Microsoft Window-based free Software for Population Genetic Analysis.
|
|
|
|
|
Zheng XY, Wolff DW, Baudracco-Arnas S, Pitrat M (1999). Development and utility of cleaved amplified polymorphic sequences (CAPS) and restriction fragment length polymorphism (RFLPs) linked to the Fom-2 Fusarium wilt resistance gene in melon (Cucumis melo). Theor. Appl. Genet. 99:453-463.
Crossref
|
|