ABSTRACT
Ensete (Ensete ventricosum) is an important perennial species in Ethiopia. It is used for food and fiber. The mode of propagation is asexual and sexual. Asexual form of reproduction is by natural and anthropogenic induced sucker. This research aims to investigate and measure the methylation diversity of natural and anthropogenic induced sucker forming Ensete. Twenty-seven individuals from Hawassa University research collection were used to identify using Amplified Fragment Length Polymorphisms (AFLPs) and Methylation-Sensitive Amplification Length Polymorphism (MS-AFLP).technology. The average values of Nei's genetic diversity (Hj) were 0.189 and 0.110 for natural sucker forming and anthropogenic induced sucker forming, respectively by AFLPs analysis. With MS-AFLP analysis, this value reduced to 0.145 for natural sucker forming Ensete but increased to 0.172 for anthropogenic sucker inducing populations. The AFLP Shannon information index (Ho) was 0.313 and 0.162 for natural and anthropogenic sucker forming Ensete, respectively. However, this value was reduced to 0.238 for natural sucker forming and 0.252 for anthropogenic sucker formed by the MS-AFLP analysis. UPGMA tree, structure analysis and principal coordinate analysis (PCoA) showed the two populations clustered separately. AMOVA revealed 24% of the genetic differentiation (Fst) occurred among populations. Gene flow (Nm) was limited among all populations. This concludes that AFLP did not show differentiation among populations; however MS-AFLP clearly showed the differentiation of populations which is an indication of epigenetic diversity but limited to developmental stages of Ensete.
Key words: Methylation, Ensete ventricosum, sucker, Amplified Fragment Length Polymorphisms (AFLPs), Methylation-Sensitive Amplification Length Polymorphism (MS-AFLP).
Abbreviation:
NE, Anthropogenic sucker inducing; ET, Natural sucker inducing; EB, sucker on the dead leaves; MB, Emerging bud; ML, emerged young shoot; D, seedling plant 1 to 9; M, Entada mother; K, Koba; G, Ganticha; A, Addo; AB, Abebe; W, Wild.
Ensete is the staple food crop in Ethiopia where, 20 million people depend on for food, feed, medicine, fiber and ornament (Pijls et al., 1995). Improving the different qualitative and quantitative traits of this crop is important to increase food security and diversity. For this, employing molecular breeding tools is an efficient approach.
The diversity of Ensete is confirmed by morphological and ethno botanical studies (Birmeta et al., 2002, 2004; Negash et al., 2002; Olango et al., 2015; Tesfaye and Lüdders, 2003; Tobiaw and Bekele, 2011; Yemataw et al., 2018). The mode of reproduction in Ensete is also different where wild Ensete has sexual propagation method by forming flower and seed, which is a unique resource for its possibility of retaining genetic recombination (Olango et al., 2015). However, cultivated Ensete population reproduces by sucker formation. This sucker formation is natural or anthropogenic induced, and most of the cultivated ensete comprised of natural sucker forming (ET) and anthropogenic sucker inducing (NE). The majority of cultivated Ensete populations which are used as source of food are NE type. However, cultivated Enset with limited use for food are ET type. Furthermore, ET and NE type of Ensete have contrasting propagation system. ET is propagated exclusively by vegetative propagation. However, NE type of Ensete is manipulated by anthropogenic activity to form multiple sucker formation. If the NE type is left till it finishes its life cycle, the plant will form a flower, the basis for sexual reproduction and seed formation.
Over the past decades, many researchers employed molecular markers such as Random Amplified Polymorphic DNA (RAPD) (Birmeta et al., 2002), Amplified Fragment Length Polymorphism (AFLP) (Negash et al., 2002), Inter Simple Sequence Repeats (ISSRs) (Tobiaw and Bekele, 2011); Single Sequence repeats (SSR) (Olango et al., 2015) and Single Nucleotides Polymorphism (SNP) (Yemataw et al., 2018) to study the genetic diversity of Ensete. Molecular markers were able to give us the genetic diversity, richness, and differentiation without the effect of environmental variation. However, above employed markers studies gave little information about the differentiation of natural and anthropogenic sucker forming of Ensete.
Epigenetic alters phenotypes of a living organism by changing the morphology and biological molecules (Broeck et al., 2018; Fournier-Level et al., 2011; Fray and Zhong, 2015; Fresnedo-Ramírez et al., 2017; Kaeppler et al., 2000; Wang et al., 2016). However, these changes are not the result of the change in DNA sequence (Dhar et al., 2019). This phenomenon challenged the traditional dogma of phenotypes controlled by DNA only. Thus currently it becomes clear that not only DNA but also epigenetic is affecting the variation and diversity of living organisms.
One of the most studied parts of the epigenetic effect is the change which happens because of DNA Methylation. Methylation-Sensitive Amplified Polymorphism (MSAP) is one the technique which is used to study DNA Methylation in the living organism (FulneÄek and KovaÅ™ík, 2014; Labra et al., 2002a, b, 2004; Pérez-Figueroa, 2013). Reyna-Lopez is the first to use this technique in Fungi (FulneÄek and KovaÅ™ík, 2014). However, after his result showing the different pattern of Methylation in clonally propagated fungi, the approach has been used by many researchers in the model and cultivated plants (Dowen et al., 2012; Fournier-Level et al., 2011; Kashino-Fujii et al., 2018; Suter and Widmer, 2013; Wang et al., 2016). Furthermore, the technique is widely applied not only to study diversity but also to understand the ecological and evolutionary aspect of the species (Broeck et al., 2018; Dowen et al., 2012). Currently, the technique is used to understand the population epigenetic diversity of living organisms. For example there is clear Methylation diversity of barley (Chwialkowska et al., 2019), Arabidopsis (Kawakatsu et al., 2016) rice (Wang et al., 2016), sorghum (Rosati et al., 2019), wheat (Shaked et al., 2001), almond (Fresnedo-Ramírez et al., 2017), grape (Fournier-Level et al., 2011), tomato (Fray and Zhong, 2015). This analysis clarified that epigenetic is important in population differentiation and influencing morphological changes. Implying epigenetic effect is an important factor for phenotypic variation. Many studies clarified DNA Methylation is the common phenomenon of a natural population which can be a potential force for the evolutionary process in living organisms. This indicates understanding the epigenetic effect is important for understanding the evolutionary and ecological explanation of population diversity. Thus, for proper management of Ensete breeding, it is necessary to evaluate the local genetic/epigenetic structure in these populations. Unfortunately, there is no information about the extent and partitioning of genetic and epigenetic diversity in Ensete populations on a local scale, as well as the genetic/epigenetic relationships between natural sucker forming (ET) and anthropogenic induced sucker formation (NE) populations of Ensete.
The major objective of this research is to provide preliminary data to determine if there is DNA Methylation differences between ET and NE Ensete trees. This study focused on dynamic differential global Methylation patterns between trees having different propagation systems. The specific objectives of the research are: (1) to check the genetic and epigenetic diversity of branching and non-branching type of Ensete, and (2) to analyze the population structure of the two types of Ensete genotypes based on their genetic and epigenetic diversity.
Study species and sampling sites
Genomic DNA was collected from emerging sucker on dead leaf (EB), emerging bud (MB), emerging shoot (ML), shoot of young plants (D), and young shoot of matured mother plants of ET ( M1, M2 , M3, HU), and shoot of matured NE plant (Ado, Ganticha, Koba, Abebe, and Wild) from the field established in Hawassa University (Figure 1). The samples consisted of two types of sucker formation: Mainly natural sucker forming-branching (ET) and anthropogenic induced sucker formation-non-branching (NE) form of Ensete (Figure 1). A total of 27 individuals' genomic DNA is extracted with the Qiagen DNeasy Plant Mini kit (Qiagen, Valencia, CA, USA).following the manufacture’s protocol.
AFLP and MS-AFLP protocol
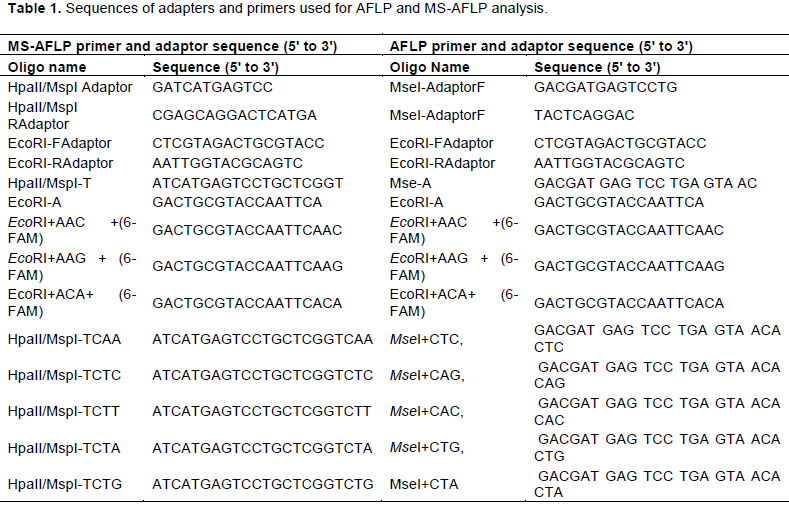
AFLP primer combinations using EcoRI+AAC (6-FAM), EcoRI+ AAG (6-FAM), and EcoRI +ACA (6-FAM) labeled primers with MseI + CTC, MseI + CAG, MseI + CAC, MseI + CTG, and MseI + CTA unlabeled primers were selected from the literature (Trebbi et al., 2019). For MS-AFLP, the MseI with MspI and HpaII isoschizomers (restricting CCGG) was replaced with different sensitivities to cytosine Methylation. The selective primers were MspI/HpaII +TCAA, MspI/ HpaII +TCTC, MspI / HpaII +TCTT and MspI/ HpaII +TCTG unlabeled primers. The primers were tested for polymorphism using two Ensete accessions (M1 and Ganticha). Following the polymorphism screening, fifteen and 9 primer combinations were selected for AFLP and MS-AFLP analysis, respectively (Table 1). Thus those polymorphic primers were used to screen all samples' genomic DNA for AFLP and MS-AFLP protocol to ensure reliable scoring of fragments. AFLP analysis was carried out using a modified protocol described by Blignaut et al. (2013), Schulz et al. (2013), Vos et al. (1995) and Vuylsteke et al. (2007). The detailed AFLP and MS-AFLP analysis were carried out with a volume of 40 µl, containing 1 µl of DNA sample (500 ng), 0.12 µl of enzyme EcoRI (Thermo Scientific TM), 0.1 µl MseI, and 0.2 µl HpaII/MspI (Thermo Scientific TM). The reaction is buffered with 4 µl Tango Restriction Buffer (Thermo Scientific TM) adjusted to the final volume of 40 with molecular grade water. The restricted DNA is incubated for 3 h at 37°C. The restricted fragment is ligated with 1 µl of T4 ligase (Invitrogen), 1 µl of ligation buffer, 1 µl of EcoRI adaptor (Thermo Scientific TM), 1 µl of MseI/HpaII/MspI adaptor (Thermo Scientific TM) and 14 µl of molecular grade water. These restriction-ligation mixtures were incubated at 37°C for 2 h and then denatured at 70°C for 15 min. The pre-selective amplification reactions were prepared in a final volume of 20 µl containing 5 µl of diluted the restricted-ligated DNA in to 1:9 dilution factor; 0.25 µl of polymerase (Invitrogen); 2 µl PCR buffer-MgCl2 (10 mM); 0.4 µl dNTPs (10 mM); 0.6 µl MgCl2 (50 mM) and 7.5 µl of water. The pre-selective PCR reactions were then amplified in a DNA thermo cycler programmed under the following conditions: 72°C for 2 min (1 cycle); 94°C for 30 s., 56°C for 1 min, 72°C for 2 min (30 cycles) and 72°C for 30 min (1 cycle). The pre-selective amplification products were then visualized on a 1.5% agarose gel. The selective amplification reactions were prepared in a final volume of 10 µl containing 2.5 µl of template DNA from pre-selective PCR, 0.625 µl of PrimerEcoRI-FAM+3 (10mM), primer MspI+3/MpaII+3/MseI+3 selective primer set (10 µM, Thermo Scientific TM), 12.5 µl of Taq polymerase (5 U/µl) adjusted to final volume of 10 with water (nuclease-free). The selective PCR reactions were then amplified in a DNA thermo cycler programmed as follows: 1 cycle at 95°C for 15 min, 95°C for 30 s, 66°C for 30 s and 72°C for 2 min. The annealing temperature was then lowered by 0.7°C per cycle during the first 12 cycles, and then 23 cycles were performed at 94°C for 30 s, 56°C for 30 s, and 72°C for 60 s, then at 4°C forever. The amplification selective PCR products were diluted to 1:9 dilution factors and denatured with formamide. A 500-bp Liz standard marker was used to estimate the molecular size of the fragments. The mixture is analyzed using the NMBU University GENE Center facility for fragment analysis on an ABI3130XL.
Data analysis
To score AFLP and MS-AFLP fragments, the gene-mapper software was used. We manually set bin widths using the graphic interface with the minimum relative fluorescence units for band identification at 50 bp. These parameters resulted from bands coded as "1" or "0" for present and absent, respectively. Throughout we use "locus" to indicate a specific fragment size in the AFLP and MS-AFLP results. "Haplotypes" was use to indicate the binary variable positions (dominant genotypes) for each individual’s collection of AFLP loci, and "epigenotype" to indicate the collection of binary variable positions of MS-AFLP loci. Both AFLP and MS-AFLP data were analyzed using GenALex version 6.5 and POPGENE version 1.31 software. The assumption of population structure was tested for both genetic and epigenetic loci by calculating standard population genetics statistics within and among populations using GenALex 6.5. GenALex was also used to estimate genetic and epigenetic differentiation using hierarchical analysis of molecular variance (AMOVA) to determine if there was structure associated with the population. This analysis assessed structured genetic or epigenetic differences by comparing variation in marker profiles (AFLP or MS-AFLP) among populations (ФRT) and within populations (ФPR). 9,999 permutations was used to estimate statistical significance and an initial alpha of 0.05.
The dendrogram was constructed based on Neil's genetic distance using the unweighted pair group method with arithmetic average (UPGMA). The partitioning of total genetic diversity (HT) into within (HS) and among (DST) accession components was examined using Nei1’s genetic diversity statistics (HT = HS + DST; GST = DST/HT).
It was initially assumed that each population is clonally propagated, thus little diversity and no epigenetic variation is expected. Using the resulting data, Bayesian clustering was performed using Structure v.2.3.4 to identify how many different groups are represented among the individuals. Structure estimates the number of groups (k) present among individuals and assigns individuals to each k using Bayesian modeling. Five populations (k=1-5) were tested which is three populations more than the maximum anticipated based on the sampling, with five independent runs at each k. We used both the log probability of observing the data (ln Pr (x|k)) method of Structure and Delta k (Earl, 2012; Evanno et al., 2005; Falush et al., 2003) which determines the number of populations that best fit the data. Clustering was performed with the admixture model, 50,000 burn-in steps, 100,000 post-burn-in steps, and correlated allele frequencies were allowed. Individuals were assigned to groups based on the highest q-value.
Phonotypical characterization of natural sucker forming and anthropogenic induced sucker forming Ensete established at Hawassa University research field
Under the normal growth conditions, there is a notable phenotypic difference observed between ET (the natural sucker forming) and Anthropogenic induced sucker forming (Ado, Ganticha, Koba, Wild type and Abebe) Ensete (Figure 1). The growth pattern of the two types of Ensete was evaluated in the established garden. The morphology show a difference pattern of growth which shows the emerging of sucker on dead leaves of the matured ET type of Ensete. However, this process is completely absent in the common cultivated Ensete types (Ado, Ganticha, Koba, and wild). These indicated that natural sucker formation is absent in the common group of Ensete while maintained in the Entada accessions. Observation of Ensete population for more than ten years (data not presented) confirmed three types of multiplication in the sampled genotypes. The first multiplication method is exclusive vegetative. This is observed only by ET and Abebe accessions. The second multiplication system is anthropogenic induced sucker formation which is observed among cultivated Ensete like Ado, Ganticha and Koba. The third multiplication system is exclusively sexual by forming seed as represented by Wild (W) type Ensete.
Polymorphism of AFLP and MS-AFLP data
The AFLP yielded a total of 3745 bands ranging from 41 to 400 bands per primer combinations (Table 2a). All primers produced polymorphic bands for both populations (Table 3). The effective number of allele ranged from 1.78 to 1.927. This value varies among ET and NE populations. The number of privet bands for NE and ET is 101 and 2051, respectively (Table 3). Unbiased haploid genetic diversity for each population also varied (0.198 and. 0.146 for ET and NE, respectively).
The MS-AFLP analysis also showed significant polymorphism by each of the nine primer combinations. The total number of bands generated by the MS-AFLP was 2846 (Table 2b). The number of bands per primer combination ranged from 14 to 319. Polymorphic bands ranged from 14 to 226. The effective number of alleles ranged from 1.585 to 2.0. The number of privets bands was 835 and 479 for ET and NE, respectively. Unbiased haploid genetic diversity for each population also varied with a value of 0.151 and. 0.229 for ET and NE, respectively. This showed an increased number of privet bands for NE compared to the AFLP analysis.
Population differentiation
Total genetic diversity per primer combination ranged from 0.145 to 0.208 for AFLP analysis. However, the genetic diversity within (HS) the population ranged from 0.135 to 0.191 indicated higher diversity within the population than among the population (Dst) which ranged from 0.011 to 0.017. However, the differentiation index (GST) ranged from 0.045 to 0.104. This value indicates lower differentiation among the population. However, for MS-AFLP, total genetic diversity (HT) ranged from 0.13 to 0.265, while the genetic diversity within (HS) the population ranged from 0.117 to 0.217. The Dst value ranged from 0.013 to 0.071, indicating low diversity. However, the differentiation index (GST) ranged from 0.09 to 0.339. The relative higher differentiation index value by the MS-AFLP analysis indicated the moderate differentiation of the population.
Analysis of molecular variance revealed no genetic differentiation among the population in the AFLP analysis but significantly higher differentiation among the population by the MS-AFLP analysis ((Φ=0.25, P = 0.001). The result of this analysis indicated that 24% of molecular variance was present among the two Ensete types, whereas 76% of the molecular variation was within the samples in the population (Table 4). This data clearly demonstrated that the presence of cytosine Methylation increased the differentiation of the population.
Cluster analysis
To further determine the genetic relationships among the two populations, UPGMA clustering was carried out using Nei’s unbiased genetic distance matrix. The dendrogram failed to revealed inter-population relationships by AFLP analysis (data is not presented). In the MS-AFLP analysis two groups (A and B) were separated by the MS-AFLP analysis. Eighteen individuals were clustered in one group (A) and nine individuals clustered into another group (B). Further, sub-group A-I and A-II are composed of 9 individual from which exclusively consisted of samples of sucker on dead plant (MB, MB2 and MB3), emerged bud (MS1, MS2 and MS3), and emerged young shoot (ML1, ML2 and ML) type ET and daughter plants (D12, D13, D17, D20, D21, D22, D37, D38 and D39) of ET mother plant. And sub-group B-I is composed of four individual of cultivated Ensete of NE type (Ado (A), Ganticha (G), Koba (K), and Wild (W)), whereas sub-group B-II is composed of the mother plants of ET (M1, M2, M 3 and HU). The exception is one individual (Abebe (AB)) which is grouped as the outlier in B-II. This implies that the MS-AFLP analysis distance is greater than the AFLP analysis confirming an important role for DNA Methylation in the Ensete epigenome since the differences between two types of Ensete are enhanced.
Structure analysis
The AFLP procedure did not cluster the population significantly (data not presented). For the MS-AFLP procedure, the structure of ET and NE was analyzed with no apriori information, using the Structure software. The result showed a clear peak (ΔK = 19.13) at the K value of 2 (Figure 2b to d) based on likelihood plots of the models, the stability of grouping patterns across different runs, and germplasm information. Group 1 had eighteen individuals while group 2 is composed of nine individuals. Group 1 is composed of the young plant from Entada (D1 to D9), a sucker from dead leaves (EB), emerging bud (MB), and emerged young shoot (ML). While group two are composed of all the anthropogenic sucker inducing plants (Ado, Ganticha, Koba), the mother plant of Entada (M1, M2, M3, HU and Abebe), and wild type which reproduced by seed formation. The result indicated that 78.8% from the ET group are clustered in the same group while 21.3% are grouped in the second group. However, 99.65 of the NE are grouped in the second group. The average distance between the individuals within the cluster was 0.12 and 0.22, for cluster 1 and cluster 2, respectively. The mean Fst value for each cluster is 0.56 and 0.11, for cluster 1 and cluster 2, respectively. The mean α value is 0.05, 0.04, 0.03 and 0.02 for K = 2, 3, 4 and 5, respectively, showing significant difference between each cluster.
Principal coordinate analysis
The genetic relationship among the studied populations was also visualized by performing PCoA based both on the AFLP and MS-AFLP data (Figure 3). For AFLP only, the first two components accounted for 14% (7.5 and 6.5%) of variation observed in the populations. The two estimated population intersected and shared a large part of the ellipse area. However, in MS-AFLP analysis only the first two components accounted for 34.3% (19.7 and 12.6%) of variation observed in the populations. Where the major and minor axes, which show the dispersion degree of the population, indicates the dispersion is higher even though some level of overlap among the populations. A similar pattern of clustering is revealed by the PCoa analysis and UPGMA dendrogram (Figure 2a and Figure 3).
This study offers an analysis of the genetic and epigenetic diversity and population structure of Ensete ventricosum in Ethiopia. Both MS-AFLP and AFLP have been proven to be valuable for the determination of genetic and epigenetic diversity among the collected genotypes in Hawassa University research field.
MS-AFLP and AFLP profiles were generated from Ensete genomic DNA extracted from plant tissue of Ado, Ganticha, Koba, Abebe, Wild type and Entada which represent the mother plants, a sucker from the dead leaves, emerging bud, emerged shoot and separated seedlings from the same Entada plant (Figure 1). The quality of AFLP and MS-AFLP was good; as a result, comparing the DNA profiles of the different tissues and plants was possible for comparing the genetic and epigenetic patterns in the selected plants.
Restriction of genomic DNA with EcoRI/HpaII/MspI creates three groups of fragments (Vos et al., 1995; Vuylsteke et al., 2007). However, the target was to measure the fragments which are preferentially amplified by EcorI-/MspI/HpaII selective primers. This lowered the number of amplified fragments in MS-AFLP analysis than the AFLP analysis.
As a result, the following four classes of fragments originated: Un-methylated, hemi-methylated internally, hemi-methylated externally and fully methylated (Schulz et al., 2013). Conservation of the MseI site indicates the genetic similarity of the germplasm while variation indicates the genetic diversity which can be quantified using GST, Amova, Nei genetic diversity and structure software (Pérez-Figueroa, 2013). The fifteen primer combinations resulted in more than 3000 loci with higher polymorphic bands. However, the AFLP polymorphic loci were not able to differentiate the two populations into natural sucker forming (ET) and anthropogenic induced sucker forming (NE) groups. There are many polymorphic bands as a result of EcorI/MseI restriction analysis. A similar result has been reported by different author using different population AFLP (Negash et al., 2002). Even if the research was done using a different collection of Ensete from different agro ecology, there is moderate genetic diversity within the population than among the population of Ensete. Depending on their population determination little or no population differentiation has been exhibited. This indicates the clones are the result of cross-pollination.
Yet, with nine pairs of primer combinations, the MS-AFLP analysis revealed more polymorphic bands in the population and significant differentiation of the two populations. This might be the result of fragments which are methylated with their internal cytosine and externals cytosine or fully Methylation as the result of the two schizomeric restriction enzymes (HpaII /MspI). This indicates that there is some level of epigenetic variation exhibited by the population. Furthermore, the small percentage of polymorphism (0.145 and 0.229 for ET and NE, respectively) found in the MS-AFLP analyses shed important information about the Methylation pattern and the presence of independent allele as a result of Methylation in the two populations (Michalakis and Excoffier, 1996). This indicates the presence of a different pattern of Methylation on the natural and anthropogenic induced sucker forming populations. Such a high level of diversity indicates the presence of high epigenetic diversity.
Changes in Methylation patterns occur by de novo Methylation which is catalyzed by Methyl transferase enzyme (Lyko, 2017). In this result, a different pattern of Methylation where the two populations exhibited differently was observed. The pattern of DNA Methylation is exhibited by the different number of unique fragments in the MS-AFLP analysis. For example, an increased number of privet bands (835) were exhibited in the ET population while less number (479) was found in the NE populations. This indicates that de-methylation of the NE population which might have happened due to the failure in maintaining the Methylation pattern through DNA replication. This might have created a different number of unique bands by MS-AFLP. The absence or disappearance of bands in the NE and ET population might be attributed to the status of the Methylation sites which need to be elucidated further.
The presence of more effectively amplified fragments by the AFLP showed a higher frequency in the ET populations. The high number of the fragment in the ET population might reveal the de-methylation of the genome sequence in the population while Methylation in the NE populations made it inaccessible for the digestion by EcoRI/MseI enzyme combinations. De-Methylation further is supported by the increased fragments number in the ET population than the NE population by the HpaII/MspI enzyme combinations that indicate the effect of the de-methylation events occurring during the different developmental stages of ET ensete.
The overall explanation of the data reflects the presence of Methylation and De-methylation in the NE and ET population which occur in Ensete population. This explanation can be supported by the fact that cytosine Methylation is a well-known phenomenon in plants as it is observed by different plant species (Broeck et al., 2018; Dowen et al., 2012; Fresnedo-Ramírez et al., 2017). Methylation affects the gene expression which in turn affects plant adaptation and productivity (Kashino-Fujii et al., 2018). Thus, de-methylation might be one aspect of differentiation in the Methylation pattern of plants at a different stage of their development. This further characterizes that the genes or the Methylation pattern of Ensete are important to unravel the developmental pattern of Ensete.
The explanation behind the variability of the two forms of Ensete (ET vs NE) based on MS-AFLP results is an indication of the selection strategy of the germplasm for vegetative reproduction. The ET form is not able to form a flower but propagate by the sucker. Thus, the absence of recombination can reduce to generate variation in the population. Thus, ET relies on epigenetic which will be the only way for the plant to exist. This could explain why the different developmental stages are the target for epigenetic variability and creating population structure.
The key evidence for the above explanation is the DNA Methylation fingerprint that was distinctly different (Φ ST = 0.25,p = 0.001) among the two forms of Ensete. This uncovers a disjunction between the global DNA Methylation pattern and the developmental state of Ensete as revealed by the structure analysis. Therefore, the difference that was observed between the Methylation fingerprints of developmental stages and genotypes could reflect the developmental stage-related phenotypic plasticity.
To date, regulation of gene expression through DNA Methylation has been described for many clonally propagated plants like grape (Fournier-Level et al., 2011), almond (Fresnedo-Ramírez et al., 2017), but not for Ensete. The Methylation patterns described in those earlier studies were generally associated with ecological contexts, developmental stages, and genotypes. In this results, there is no geographical scope where all the population is established in the same environment for more than three years. But epigenetic analysis differed between the different populations where little genetic diversity is exhibited. There are few reports with big genetic variation and the epigenetic difference in plants. The current research supports the idea of change in Methylation involved in the developmental stage of the plant which includes a natural sucker forming Ensete. This concept could open new perspectives for a better understanding of Ensete developmental biology and mechanism of sucker formation. The epigenome analysis of branching and non-branching Ensete population along with different ecology is needed. Discovering the key genes involved in the change in the branching and non-branching patter mechanism, as well as their epigenetic variation, is necessary to clarify the pattern of epigenetic variation evidenced in this study.
Results indicate that the genetic diversity of Ensete ventricosum is high based on MS-AFLP analysis. Less genetic differentiation occurred among the AFLP markers which are based on the genetic difference. This study highlights the presence of Epigenetic variation among the population which should be further elucidated. The information generated from this study will facilitate future works on the epigenetic relationship and different morphological and developmental phenomena in E. ventricosum which is one of the important crop plants in Ethiopia.
The authors have not declared any conflict of interests.
NORHED project “Controlling diseases in sweet potato and Enset in South Sudan and Ethiopia”.
NE, Anthropogenic sucker inducing; ET, Natural sucker inducing; EB, sucker on the dead leaves; MB, Emerging bud; ML, emerged young shoot; D, seedling plant 1 to 9; M, Entada mother; K, Koba; G, Ganticha; A, Addo; AB, Abebe; W, Wild.
This research was supported by NORHED project “Controlling diseases in sweet potato and Enset in South Sudan and Ethiopia”.
REFERENCES
|
Birmeta G, Nybom H, Bekele E (2002). RAPD analysis of genetic diversity among clones of the Ethiopian crop plant Ensete ventricosum. Euphytica 124(3):315-325.
Crossref
|
|
|
|
Birmeta G, Nybom H, Bekele E (2004). Distinction between wild and cultivated Enset (Ensete ventricosum) gene pools in Ethiopia using RAPD markers. Hereditas 140(2):139-148.
Crossref
|
|
|
|
|
Blignaut M, Ellis AG, Roux JL (2013). Towards a Transferable and Cost-Effective Plant AFLP Protocol. PloS One 8:e61704.
Crossref
|
|
|
|
|
Broeck AV, Cox K, Brys R, Castiglione S, Cicatelli A, Guarino F, Heinze B, Steenackers M, Vander Mijnsbrugge K (2018). Variability in DNA Methylation and Generational Plasticity in the Lombardy Poplar, a Single Genotype Worldwide Distributed Since the Eighteenth Century. Frontiers in Plant Science 9:1635
Crossref
|
|
|
|
|
Chwialkowska K, Korotko U, Kwasniewski M (2019). DNA Methylation Analysis in Barley and Other Species with Large Genomes. In Barley: Methods and Protocols, WA Harwood, ed. (New York, NY: Springer New York) pp. 253-268.
Crossref
|
|
|
|
|
Dhar MK, Sharma R, Vishal P, Kaul S (2019). Epigenetic Response of Plants to Abiotic Stress: Nature, Consequences and Applications in Breeding. In Genetic Enhancement of Crops for Tolerance to Abiotic Stress: Mechanisms and Approaches. Vol. I, Rajpal VR, Sehgal D, Kumar A, Raina SN, eds. (Cham: Springer International Publishing), pp. 53-72.
Crossref
|
|
|
|
|
Dowen RH, Pelizzola M, Schmitz RJ, Lister R, Dowen JM, Nery JR, Dixon JE, Ecker JR (2012). Widespread dynamic DNA methylation in response to biotic stress. Proceedings of the National Academy of Sciences of the United States of America 109:E2183-E2191.
Crossref
|
|
|
|
|
Earl DA (2012). Structure harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4(2):359-361.
Crossref
|
|
|
|
|
Evanno G, Regnaut S, Goudet J (2005). Detecting the number of clusters of individuals using the software Structure: a simulation study. Molecular Ecology 14(8):2611-2620.
Crossref
|
|
|
|
|
Falush D, Stephens M, and Pritchard JK (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164(4):1567-1587.
|
|
|
|
|
Fournier-Level A, Hugueney P, Verriès C, This P, Ageorges A (2011). Genetic mechanisms underlying the methylation level of anthocyanins in grape (Vitis vinifera L.). BMC Plant Biology 11:179.
Crossref
|
|
|
|
|
Fray R, Zhong S (2015). Genome-wide DNA methylation in tomato. In: Applied Plant Genomics and Biotechnology, Poltronieri P, Hong Y. eds. (Oxford: Woodhead Publishing) pp. 179-193.
Crossref
|
|
|
|
|
Fresnedo-Ramírez J, Chan HM, Parfitt DE, Crisosto CH, Gradziel TM (2017). Genome-wide DNA-(de)methylation is associated with Noninfectious Bud-failure exhibition in Almond (Prunus dulcis [Mill.] D.A.Webb). Scientific Reports 7:42686.
Crossref
|
|
|
|
|
FulneÄek J, KovaÅ™ík A (2014). How to interpret methylation sensitive amplified polymorphism (MSAP) profiles? BMC Genetics 15:2.
Crossref
|
|
|
|
|
Kaeppler SM, Kaeppler HF, Rhee Y (2000). Epigenetic aspects of somaclonal variation in plants. Plant Molecular Biology 43(2-3):179-188.
Crossref
|
|
|
|
|
Kashino-Fujii M, Yokosho K, Yamaji N, Yamane M, Saisho D, Sato K, Ma JF (2018). Retrotransposon Insertion and DNA Methylation Regulate Aluminum Tolerance in European Barley Accessions. Plant Physiology 178:716-727.
Crossref
|
|
|
|
|
Kawakatsu T, Huang SC, Jupe F, Sasaki E, Schmitz RJ, Urich MA, Castanon R, Nery JR, Barragan C, He Y (2016). Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166(2):492-505.
Crossref
|
|
|
|
|
Labra M, Ghiani A, Citterio S, Sgorbati S, Sala F, Vannini C, Ruffini-Castiglione M, Bracale M (2002a). Analysis of cytosine methylation pattern in response to water deficit in pea root tips. Plant Biology 4(6):694-699.
Crossref
|
|
|
|
|
Labra M, Vannini C, Bracale M, Sala F (2002b). Methylation changes in specific sequences in response to water deficit. Plant Biosystems 136(3):269-275.
Crossref
|
|
|
|
|
Labra M, Grassi F, Imazio S, Di Fabio T, Citterio S, Sgorbati S, Agradi E (2004). Genetic and DNA-methylation changes induced by potassium dichromate in Brassica napus L. Chemosphere 54(8):1049-1058.
Crossref
|
|
|
|
|
Lyko F (2017). The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nature Reviews Genetics 19(2):81-92
Crossref
|
|
|
|
|
Michalakis Y, Excoffier L (1996). A generic estimation of population subdivision using distances between alleles with special reference for microsatellite loci. Genetics 142(3):1061-4.
|
|
|
|
|
Negash A, Tsegaye A, van Treuren R, Visser B (2002). AFLP Analysis of Enset Clonal Diversity in South and Southwestern Ethiopia for Conservation. Crop Science 42(4):1105-1111.
Crossref
|
|
|
|
|
Olango TM, Tesfaye B, Pagnotta MA, Pè ME, Catellani M (2015). Development of SSR markers and genetic diversity analysis in enset (Ensete ventricosum (Welw.) Cheesman), an orphan food security crop from Southern Ethiopia. BMC Genetics 16:98
Crossref
|
|
|
|
|
Pérez-Figueroa A (2013). msap: a tool for the statistical analysis of methylation-sensitive amplified polymorphism data. Molecular Ecology Resources 13(3):522-527.
Crossref
|
|
|
|
|
Pijls LJ, Timmer,AM, Wolde-Gebriel Z, West CE (1995). Cultivation, preparation and consumption of ensete (Ensete ventricosum) in Ethiopia. Journal of the Science of Food and Agriculture 67(1):1-11.
Crossref
|
|
|
|
|
Rosati VC, Quinn AA, Fromhold SM, Gleadow R, Blomstedt CK (2019). Investigation into the role of DNA methylation in cyanogenesis in sorghum (Sorghum bicolor L. Moench). Plant Growth Regulation 88(1):1-13.
Crossref
|
|
|
|
|
Schulz B, Eckstein RL, Durka W (2013). Scoring and analysis of methylation-sensitive amplification polymorphisms for epigenetic population studies. Molecular Ecology Resources 13:642-653.
Crossref
|
|
|
|
|
Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA (2001). Sequence Elimination and Cytosine Methylation Are Rapid and Reproducible Responses of the Genome to Wide Hybridization and Allopolyploidy in Wheat. Plant Cell 13(8):1749-59
Crossref
|
|
|
|
|
Suter L, Widmer A (2013). Environmental heat and salt stress induce transgenerational phenotypic changes in Arabidopsis thaliana. PloS One 8:e60364.
Crossref
|
|
|
|
|
Tesfaye B, Lüdders P (2003). Diversity and distribution patterns of enset landraces in Sidama, Southern Ethiopia. Genetic Resources and Crop Evolution 50(4):359-371.
Crossref
|
|
|
|
|
Tobiaw DC, Bekele E (2011). Analysis of genetic diversity among cultivated enset (Ensete ventricosum ) populations from Essera and Kefficho, southwestern part of Ethiopia using inter simple sequence repeats (ISSRs) marker. African Journal of Biotechnology 10(70):15697-15709.
Crossref
|
|
|
|
|
Trebbi D, Ravi S, Broccanello C, Chiodi C, Stevanato P (2019). Genomic Resources and Marker-Assisted Selection in Jatropha curcas. In Jatropha, Challenges for a New Energy Crop, (Springer) pp. 145-160.
Crossref
|
|
|
|
|
Vos P, Hogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Friters A, Pot J, Paleman J, Kuiper M (1995). AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research 23(21):4407-4414.
Crossref
|
|
|
|
|
Vuylsteke M, Peleman JD, van Eijk MJ (2007). AFLP technology for DNA fingerprinting. Nature Protocols 2(6):1387-1398
Crossref
|
|
|
|
|
Wang W, Qin Q, Sun F, Wang Y, Xu D, Li Z, Fu B (2016). Genome-Wide Differences in DNA Methylation Changes in Two Contrasting Rice Genotypes in Response to Drought Conditions. Frontiers in Plant Science 7(507):1-13.
Crossref
|
|
|
|
|
Yemataw Z, Muzemil S, Ambachew D, Tripathi L, TesfayeK, Chala A, Farbos A, O'Neill P, Moore K, Grant M (2018). Genome sequence data from 17 accessions of Ensete ventricosum, a staple food crop for millions in Ethiopia. Data in Brief 18:285-293.
Crossref
|
|