ABSTRACT
Carboxypeptidase A (CPAs) are a well-studied group of zinc-containing exopeptidases that facilitate the breakdown of proteins and peptides during metabolism. Carboxypeptidase A is typically produced in mammalian pancreatic, brain and other tissues. A new gene encoding carboxypeptidase A in the prokaryote Bacillus pumilus was amplified by polymerase chain reaction (PCR), ligated into the shuttle vector pMA5, and cloned in a GRAS bacteria-Bacillus subtilis 168 host. This gene sequence contained a 1621 bp open reading frame that encodes a protein of 540 amino acids. The optimum pH and temperature for enzyme activity were 7.5 and 50°C, respectively. The enzyme was quite stable at neutral pH and maintained about 65% activity following a 24 h incubation at 40°C. The Km of this CPA was 0.1 mM, much higher than in mammalian species. Glycerol, ammonium sulfate, and sodium citrate improved enzyme activity under optimal culture condition. The carboxypeptidase activity in recombinant B. subtilis 168 reached a maximum of 179 U ml-1 in a 5 L fermentator when cultured on improved medium. The over expression of carboxypeptidase A in Bacillus subtilis has commercial applications.
Key words: Bacillus pumilus, Bacillus subtilis 168, over-expression, orthogonal arrays, carboxypeptidase A, metallocarboxypeptidase.
Carboxypeptidases (CPs) catalyze the release of C-terminal amino acids from proteins and peptides (Sebastian Tanco et al., 2013). CPs serve many important functions in a variety of organisms since originally isolated from bovine pancreatic tissue in 1929 (Suwen et al., 2002). Some non-digestive zinc carboxypeptidases are involved in hormone and neuropeptide processing, bioactive peptide activation or inactivation, or functional modulation of regulatory proteins (Joshi et al., 1999). Thus, CPs are widely used
in the pharmaceutical, food and other industries. For example, they are used to remove ochratoxin A (Abrunhosa et al., 2007) and as a debitterizing reagent in the hydrolysis of soybean protein (Fang et al., 2005). Carboxypeptidases are classified into several types, carboxypeptidases that have a stronger preference for those amino acids containing aromatic or branched hydrocarbon chains are called carboxypeptidase A (A for aromatic/aliphatic), carboxypeptidases that cleave positively charged amino acids (arginine, lysine) are called carboxypeptidase B (B for basic), metallo-carboxypeptidase that cleaves a C-terminal glutamate from the peptide N-acetyl-L-aspartyl-L-glutamate is called "glutamate carboxypeptidase´´.
Carboxypeptidase A (CPA), a type of metallocarboxypeptidase, preferentially removes C-terminal amino acid residues in the presence of aromatic or branched aliphatic side chains with the aid of a Zn2+ ion (Austin et al., 2011). CPA has been widely studied, including the structures of the substrate-binding sites and the mechanism of catalysis (Vertesi et al., 1999). Many characteristics of CPA have been reported in literature (Kumar et al., 2014; Elena et al., 2007; Li and Solomon, 1997; Kazuhisa anda Kamisosoyama, 2000). Proposed commercial applications of CPA include its use in the hydrolysis of cheese whey protein and other protein hydrolysates. However, commercial production of CPA involves its isolation from pancreatic tissue which limits production (Lyons et al., 2008). CPA has been studied in numerous mammalian species including human, rat, bovine, swine, goat and buffalo. CPA has been studied in avian species (Kazuhisa and Kamisosoyama, 2000). CPA production by the fungi Aspergillus niger and Metarhizium anisopliae have been reported by Abrunhosa and Venancio (2007) and Austin et al. (2011), respectively. While most studies have focused on CPA characterization and applications, few studies have focused on improving the production of CPA. Heterologous expression and production of CPA in engineered strains of bacteria could increase the production of CPA.
Bacillus pumilus strains are highly resistant to UV radiation and hydrogen peroxide, making them good model bacteria for studying of industrial production strains. Nagamori et al. (1991) examined the enzymatic properties of dipeptidylcarboxypeptidase (DCP) isolated from B. pumilus; they found that the presence of chloride ion is essential for the hydrolysis, enzyme is readily inhibited by Ethylenediaminetetraacetic acid (EDTA) but restored by Co2+, Mn2+, and Zn2+. However, there is no information on the characterization of carboxypeptidase A from B. pumilus, or from other prokaryotic bacteria. Bacillus species have been major workhorse industrial microorganisms with roles in applied microbiology (Schallmey et al., 2004). Engineered strains of B. subtilis are attractive as industrial organisms due to their rapid growth rate that result in short fermentation cycle times and their capacity to secrete proteins into the extracellular medium (Gao et al., 2013).
In this study B. subtilis 168 was utilized as a host for the heterologous expression of a gene encoding carboxypeptidase A from B. pumilus. The CPA was also
characterized.
Strains, plasmids, and chemicals
B. pumilus ML413, E.coli JM109, B. subtilis 168 and vector pMA5 were preserved in our laboratory. Plasmid pMA5 and B. subtilis168 were used as the expression vector and host cell, respectively. The vector pMD18-T, restriction enzymes, T4 DNA ligase, and ExTaq DNA polymerase were purchased from TaKaRa Bio Co., Ltd. (Dalian, China). The Mini Chromosome Rapid Isolation Kit, Mini Plasmid Rapid Isolation Kit, and Mini DNA Rapid Purification kit were obtained from Generay Biotech Co., Ltd. (Shanghai, China). AKTA prime plus and Butyl Sepharose HP columns were purchased from GE Healthcare, Inc. (Little Chalfont, UK). PCR primers and Bradford reagent kits were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). Hippuryl-L-phenylalanine was purchased from Sigma-Aldrich Co., Ltd. (St. Lous, MO, USA). All other chemicals were of analytical grade and purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China).
Medium and culture conditions
B. pumilus, E.coli JM109 and B. subtilis 168 were grown at 37°C in Luria-Bertani (LB) medium during the construction of strains. Ampicillin (100 μg/ml) or kanamycin (50 μg/ml) were added to the growth medium as required. The basal fermentation medium for producing CPA was Terrific Broth (TB) medium (Gao et al., 2014) containing (g/L): yeast extract, 24; tryptone, 12; K2HPO4, 9.4; KH2PO4, 2.2; glycerol, 5, at pH 7.2. Optimization of medium was carried out based (by completing the TB medium with carbon sources, nitrogen sources, and inorganic salts) on the TB medium. Liquid cultures were incubated at 37°C at 200 rpm.
Cloning and construction of recombinant plasmid pMA5-cpa
Total genomic DNA of B. pumilus ML413 was extracted using a mini chromosome rapid isolation kit. The cpa gene was amplified from total chromosomal DNA isolated from B. pumilus ML413 by polymerase chain reaction (PCR) using ExTaq DNA polymerase, with cpa forward (5’-CGGGATCCCGATGTCCAAACAGAAAAGCACCATT-3’) and reverse (5’-CACGCGTCTTAGTGGTGGTGGTGGTGGTGTTTTATACCTAACGT-3’) primers (underlined sequence = restriction sites), which were designed using the hypothetical (yes) M14 family carboxypeptidase gene of B. pumilus SAFR-032 (accession number CP000813.1) available from the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/). The restriction sites of the forward and reverse primers were BamHI and MluI, respectively. Procedures for PCR were carried out as: 94°C for 5 min followed by 34 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 2 min. PCR amplicons were checked by agarose electrophoresis and purified from the gel using the Mini DNA rapid purification kit. The purified fragment was ligated directly into the cloning vector pMD18-T, which was then used to transform chemically competent E. coli strain JM109. The presence of the correct plasmid was verified using BamHI and MluI restriction digests. The digested fragment was purified and then ligated with pMA5, which had already been digested using BamHI and MluI restriction enzyme. The ligation mixture was used to transform E.coli JM109 competent cells. Restriction digestion analysis was carried out to verify the presence of the recombinant plasmid pMA5-cpa gene. Nucleotide sequence of cpa insert was analyzed by Sangon Biotech. Protein and nucleotide sequence comparisons were performed using the BLAST server available from the NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and then this recombinant plasmid was transformed into chemically competent B. subtilis 168 cells for Carboxypeptidase A production using the procedure described by Spizizen (Jia et al., 2013).
Expression of cpa in B. subtilis
Recombinant B. subtilis 168 was cultivated overnight (12 h) on selective LB medium supplemented with 50 μg/ml kanamycin. The cells were then harvested by centrifugation at 10,000 g for 10 min at 4°C, and the supernatant was used for a CPA enzyme activity assay. The precipitated pellets were washed twice with 5 ml of 50 mM Tris/HCl buffer (pH 7.5), then suspended in 5 ml of the same buffer supplemented with 50 µl of lysozyme solution and kept on ice for 2 h. The mixture was then sonicated 10 times for 2 s with 5 s cooling intervals. Cell extracts were centrifuged for 30 min at 10,000 g at 4°C in a SIGAMA 4K-15 centrifuge (Sigma-Aldrich Co., Ltd., Shanghai, China) to remove cell debris. The supernatant was used for crude enzyme activity assay, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (12% acrylamide), or stored at -20ï‚°C for further analysis.
Enzyme activity assay
Enzyme activities of the intracellular and extracellular portions of the protein were measured spectrophotometrically by recording the increase in absorbance at 254 nm that accompanies the hydrolysis of the peptide substrate hippuryl-L-phenylalanine as described by (Tardioli et al., 2003) with slight modifications. Assays were performed using a UV-1200 spectrophotometer (Pharmacia Biotech), with thermostatically controlled at 25°C. A 100 µl aliquot of enzyme solution was added to the assay solution (1.45 ml of 1 mM hippuryl-L-phenylalanine prepared in 50 mM Tris-HCl buffer, pH 7.5) supplemented with 40 µmol ZnSO4.7H2O, and the increase in absorbance was measured for 5 min. The amount of phenylalanine released by CPA action was calculated from the millimolar extinction coefficient at 254 nm (0.36 l mmol-1•cm-1), and enzymatic rate was expressed as concentration of product formed per min (µmol L-1 min-1). One unit (U) was defined as the amount of enzyme that hydrolyses 1.0 µmol of hippuryl-L-phenylalanine per min at 25°C. Protein concentration was determined by the Bradford method with bovine serum albumin as the standard (David et al., 2004).
Enzyme purification
All purifications were performed using an ÄKTA Prime Plus (GE Healthcare Bio-sciences procedures). The CPA-containing supernatant was filtered through a 0.2 µm cellulose acetate membrane (Corning Incorporated, NY, USA) and applied to a 2.5 ml HisTrap column (AmershamBioSciences) equilibrated in buffer A with 50 mM Tris-HCl (pH 7.4). Then the enzyme was eluted in a linear gradient with a change of imidazole concentration (500 mM) using a flow rate of 1.0 mL min-1. The active fractions were collected (Figure 7) and dialyzed for CPA enzyme assay or storing
at -20°C.
Enzyme characterization
Optimum pH was determined by measuring the activity of purified CPA at pH 3.0 to 10.0 (pH 3.0 to 6.5, 50 mM citrate-sodium citrate buffer; pH 6.5 to 8.0, 50 mM Tris-HCl buffer; pH 8.0 to 10.0, 50 mM glycine-NaOH buffer), while the optimum temperature was examined in 50 mM Tris-HCl buffer (pH 7.5) using the standard reaction mixture. The pH and thermal stability of CPA was determined by incubation at different pH levels at 4°C for 24 h and at various temperatures for 24 h, respectively. Effects of various compounds on enzyme activity were examined in the standard reaction mixture supplemented with various cations and EDTA at concentrations of 1 mM. Enzyme in the absence of metal ions or EDTA served as a control. Kinetic parameters were determined in 50 mM Tris-HCl buffer (pH 7.5) at 25°C by changing the concentration of the substrate. Eadie-Hofstee plots were used to calculate kinetic parameters Km and Vmax according to the enzyme reactions.
Single-factor-at-a-time experiments for medium optimization
To determine the effect of carbon, nitrogen sources and inorganic salt on enzyme activity, the growth medium was supplemented with sucrose, maltose, lactose, glucose, galactose, glycerol and soluble starch as carbon sources, ammonium chloride, ammonium sulfate, ammonium acetate and urea as nitrogen sources and sodium chloride, potassium chloride, sodium nitrate, potassium nitrate and sodium citrate as inorganic salts. Each source was used at a concentration of 0.5% (w/v). Enzyme activity was determined after 12 h of incubation at 37°C and 200 rpm (revolutions per minute).
Orthogonal array methodology for medium optimization
To examine the interactions among nutritional components of fermentation medium and to optimize their concentrations for CPA production, orthogonal arrays were used as reported by Vijayalakshmi et al. (2011). An L9 orthogonal array in three levels was used consisting of 9 different experimental trials for the medium optimization to increase CPA activity (Tables 3 and 4). The design for the L9 orthogonal arrays was developed and analyzed using “zhengjiaozhushou” Chinese software. Zhengjiaozhushou design is an independent quadratic design in that it does not contain an embedded factorial or fractional factorial design. In this design the treatment combinations are at the midpoints of edges of the process space and at the center. These designs are rotatable (or near rotatable) and require 3 levels of each factor. The designs have limited capability for orthogonal blocking compared to the central composite designs.
5 L fermentation for CPA production
The recombinant B. subtilis 168 harboring plasmid pMA5-cpa was scaled up to 2 L in optimized medium in a 5 L bioreactor (Shanghai Baoxing Bioengineering Equipment Co., ltd). Inoculum (100 ml) was added to the bioreactor. The agitation rate was set to 300 rpm at 37°C. For controlled pH cultivations, the pH was maintained at 7.2 by addition of 0.5 M NaOH and 1 M HCl solution. After 8 h incubation, the parameters like optical density of cell (OD600), enzyme activity, and protein concentration were determined at
regular intervals.
Cloning, construction of recombinant plasmid pMA5-cpa
The carboxypeptidase A (CPA) gene was successfully amplified from the genomic DNA of Bacillus pumilus ML413 strain by PCR. Electrophoresis revealed that the amplicon consisted of a 1621 bp fragment (Figure 1A). BLAST analysis showed that the gene sequence had a high level of similarity with that of several Peptidase M14 containing strains published in the NCBI database including B. pumilus strain W3 (CP011150.1) and B. pumilus strain MTCC B6033 (CP007436.1) with similarities of 99 and 98%, respectively. The PCR product was ligated with pMD18-T to create pMD18-cpa, and then digested with BamHI and MluI. The digestion product was cloned into the expression vector pMA5 under the control of the HpaII promoter, resulting in recombinant vector pMA5-Pept (Figure 1C). The recombinant plasmid was digested with BamHI and MluI to produce two major fragments with sizes of 1621 and 7500 bp (Figure 1B). This result confirmed the successful construction of the recombinant plasmid.
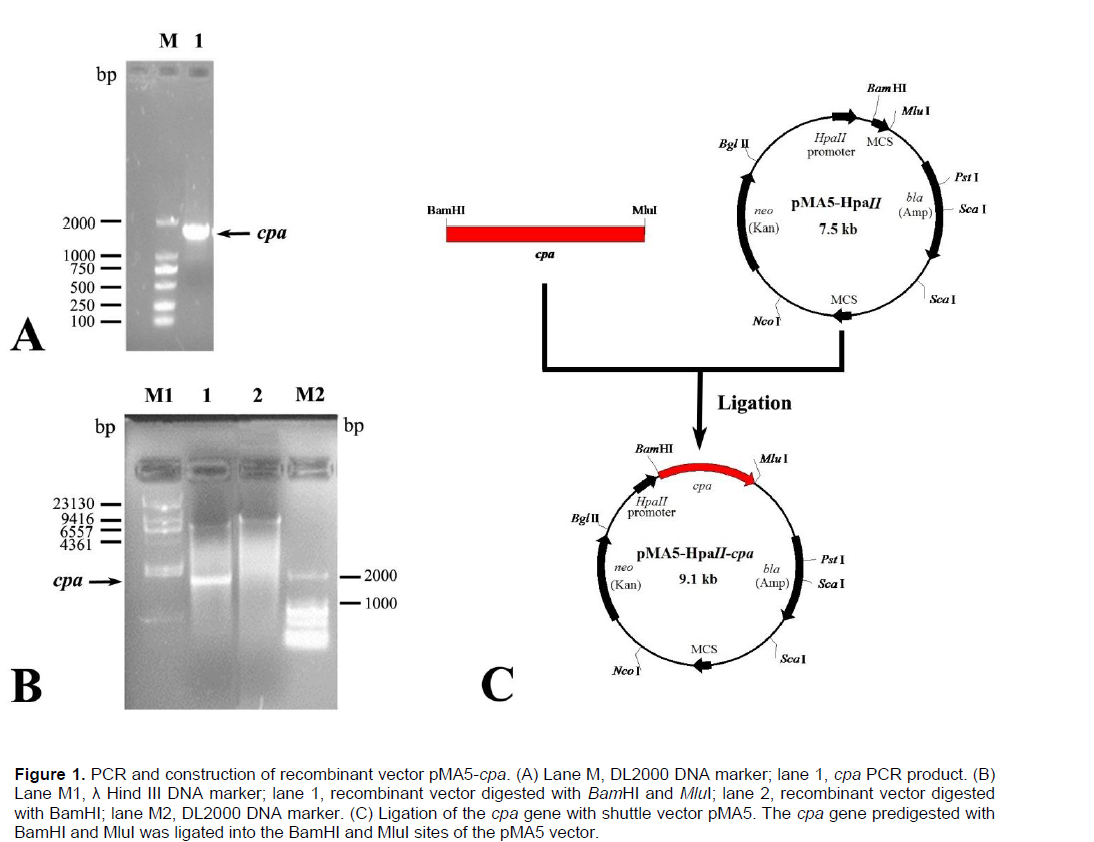
Expression of the recombinant vector in B. subtilis 168
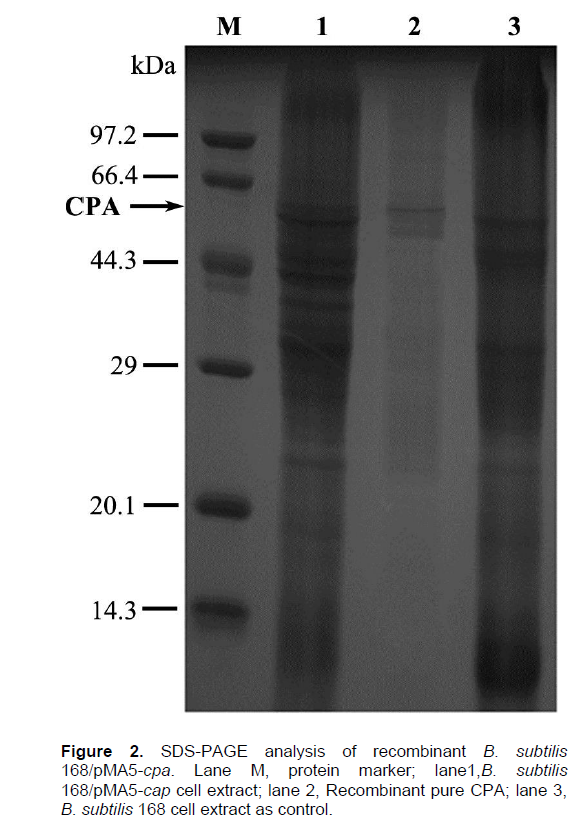
The resultant pMA5-cpa was used to transform the expression host B. subtilis 168 to construct a recombinant B. subtilis strain 168/pMA5-cpa. The recombinant B. subtilis could express CPA under the control of the HpaII promoter. For production of CPA, the recombinant B. subtilis was used to inoculate 50 ml of LB medium (pH 7.2) supplemented with kanamycin (50 µg ml-1) in a 250 ml flask, and incubated at 37°C in a rotary shaker at 200 rpm for 12 h. The recombinant B. subtilis showed intracellular and extracellular enzyme activities of 29.49 and 0 U ml-1, respectively. The total enzyme activity was substantially higher than the 7 U ml-1 of the host B. subtilis 168. A band of 59 kDa corresponding to CPA was produced by the recombinant B. subtilis (Figure 2). Unlike other mammal digestive carboxypeptidases which are activated through limited proteolysis in the duodenum (Pedro José et al., 2002), our active recombinant CPA was expressed directly. However, the recombinant CPA could not be expressed outside the host B. subtilis, while it performed as an extracellular enzyme in B. pumilus (data not shown). Gao et al. (2013) reported that TB medium was suitable for the production of enzymes by recombinant engineered strains. Hence, we cultured the recombinant B. subtilis into 50 ml TB medium using the other conditions as described for the LB medium. Intracellular and extracellular enzyme activities of CPA in TB were 45 and 0 U ml-1, respectively.
Purification of the enzyme
For affinity purification of recombinant proteins expressed in B. subtilis 168, a His tag (Tgg, Tgg, Tgg, Tgg, Tgg, Tgg) was inserted into the sequence of the reverse primer. Affinity tags are useful tools for the production of recombinant proteins because of the easier detection and purification of their fusion partners. Affinity tags may also have a beneficial impact on the yield of recombinant proteins, increase the recombinant protein’s solubility, and promote their proper folding (Bown et al., 2004). This facilitates the further study of these proteins. Recombinant CPA was purified by metal affinity chromatography, making use of a His tag at the C terminus of CPA. The SDS-PAGE of lane 2 in Figure 2 indicated that this recombinant CPA was successfully purified with a band of 59 kDa. As a result, the purified enzyme showed a specific activity of 45 U mg-1 (Table 1). The specific activity of the recombinant CPA approached the activity of commercial CPA derived from bovine pancreas (52 U mg-1) (Vertesi et al., 1999). This suggests a potential commercial use of our recombinant CPA. SDS-PAGE analysis indicated that the subunit of the enzyme had a molecular mass of about 59 kDa (Figure 2), when the gel was stained with CBB R-250.

Characterization of the Enzyme
The optimum pH values of CPA for hydrolyzing hippuryl-L-phenylalanine was pH 7.5 (Figure 3a). The optimum pH value reported here was different from values reported for carboxypeptidase from Archaeon Thermococcus sp. NA1 (pH 6) (Hyun Sook Lee et al., 2006), crayfish carboxypeptidase from Astacus fluviatilis (pH 6.5) and carboxypeptidase III from germinating triticale grains (pH 4.6) (Drzymala et al., 2009); but similar to immobilized carboxypeptidase A and carboxypeptidase A from the pancreas of the catfish Parasilurus asotus (Yoshinaka et al., 1985).
In the present study, the recombinant CPA showed maximum activity at 50°C, and then abruptly decreased at higher temperatures (Figure 3b). This was in agreement
with earlier reports of the optimum temperature of carboxypeptidase A from chicken pancreas (Kazuhisa et al., 2000). The thermal stability of our recombinant CPA is shown in Figure 3c. The recombinant CPA was stable when the temperature was less than 40°C. The activity remained at 86 and 65% at 20 and 40°C after 24 h storage, respectively. The effectiveness of pH on the stability of the CPA was examined in the range of pH 6 to 10 (Figure 3d). The recombinant CPA was found to be stable only at pH 7.5 and the activity was 98% after 24 h incubation. The influences of metal ions and EDTA on the CPA activity are as shown in Figure 4. These results indicated that the CPA enzyme was activated by Li+ (117%), Cu2+ (115%), Co2+ (110%) and Ca2+ (107%). Mg2+ had no effect on CPA activity. Fe2+ (74%) and EDTA (57%) inhibited CPA activity. Some metal ions, notably cobalt, slightly enhanced the enzyme activity of the carboxypeptidase involved in the proteolytic cleavage of the influenza haemagglutinin as reported by Garten et al. (1983).
The Km and Kcat values for recombinant CPA using hippuryl-L-phenylalanine as substrate were 0.1 mM and 63.6 s-1, respectively (as shown in Table 2). These values are similar to those of CPA from chicken pancreas, but lower than the values reported for ostrich carboxypeptidase (0.41 mM), bovine carboxypeptidase (1.75 mM), and the value reported by Cheng et al. (1999) for carboxypeptidase from the hyperthermophilic archaeon Pyrococcus furiosus (0.9 mM). These results indicate that substrate specificity of our recombinant CPA shows a higher affinity for hippuryl-L-phenylalanine when compared to that of mammals and other avian species. This suggests a potential commercial application.
Single-factor-at-a-time optimization strategy
The production of enzymes from microorganisms is strongly influenced by the composition of the medium, with carbon and nitrogen sources being the most important factors. Of the seven carbon (Figure 5a) and five nitrogen sources (Figure 5b) tested, glycerol and ammonium sulfate resulted in the highest relative activities of 197.21 and 175.85%, respectively. Lower relative activities were observed with lactose (77%) and ammonium chloride (94%). The carbon and nitrogen sources used to increase production of recombinant CPA in this study are different than those reported by (Imtiaz az et al., 2013) for the production of alkaline protease by B. subtilis, or other recombinant enzymes using B. subtilis as a host (Jia et al., 2013). The highest relative activity of carboxypeptidase (162.28%) was observed when sodium citrate was added to the culture medium as an inorganic salt source (Figure 5c). Sodium citrate was added in its simple form, so it was readily available for protein synthesis. A lower level of activity was observed when medium contained sodium nitrate or potassium nitrate. The inhibitory effects of nitrate were reported by Bhutto et al. (2011).

Optimization of enzyme activity using orthogonal array (OA) methodology
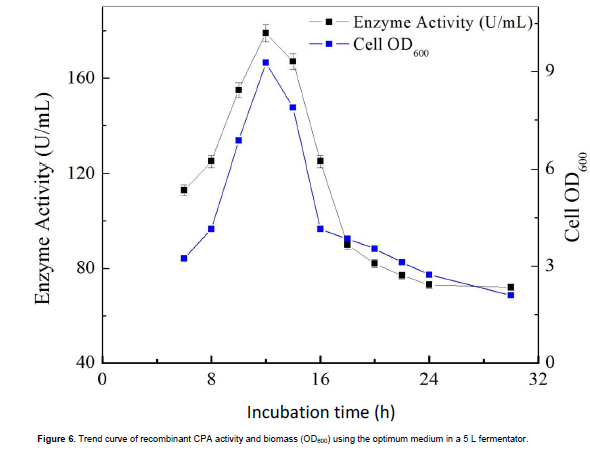
The orthogonal array methodology was used to investigate the influence of glycerol, ammonium sulfate and sodium citrate concentrations on CPA production. There are three independent variables (Glycerol, G1= 0.25%; G2 = 0.5%; G3 = 0.75%), (ammonium sulfate, As1 = 0.25%; As2 = 0.5%; As3 = 0.75%), and (sodium citrate, Sc1 = 0.25%, Sc2 = 0.5%; Sc3 = 0.75%). The order of factor effects on CPA production was found to be glycerol > sodium citrate > ammonium sulfate as shown in Table 5. The results of the optimization medium on CPA activity in 5 L fermenter are shown in Figure 6. The optimized medium contained yeast extract 24 g L-1, tryptone 12 g L-1, K2HPO4 9.4 g L-1, KH2PO4 2.2 g L-1, glycerol 5 g L-1, ammonium sulfate 5 g L-1 and sodium citrate 7.5 g L-1. The predicted level of CPA activity was 45 U ml-1 in a 50 ml flask culture. This increased to 179 U ml-1 of recombinant CPA after scaling up to 2 L in a lab scale bioreactor (12 h after inoculation). The optimized medium resulted in a 4-fold higher level of activity when compared to TB basal medium and 6-fold higher activity when compared to LB medium. The activity curve of the recombinant CPA in a bioreactor was similar to the curve of the OD600 (optical density, of sample measured at a wavelength of 600 nm), this could be due to the fact that the decline growth phase of the recombinant B. subtilis began after 16 h in such an abundant nitrogen source medium. Enzyme activity is generally directly related to the incubation period and metabolite concentrations as reported by Imtiaz et al. (2013).

A new carboxypeptidase A gene isolated from B. pumilus was successfully cloned into a Generally Recognized As Safe (GRAS) bacterium-B. subtilis 168. This enzyme was characterized after purification. The recombinant CPA was expressed in a mature form with a molecular weight of 59 kDa. This was in contrast to other mammalian digestive CPAs which need be activated through limited proteolysis with a scission of a ~100 residue N-terminal prosegment. Culture conditions, including incubation period, had a profound effect on the production of enzyme. Glycerol and ammonium sulfate provided the carbon and inorganic nitrogen sources, respectively. The recombinant carboxypeptidase A showed some characteristics in common with CPA isolated from other sources. The CPA has a broad specificity for C-terminal amino acid residues with aromatic or branched aliphatic . With a high activity of 179 U ml-1 in 5 L bioreactor, the recombinant CPA expressed in the GRAS bacterium-B. subtilis proves promising as a processing aid for protein hydrolysis in order to improve flavor and reduce bitterness in food protein hydrolysates.
The authors have not declared any conflict of interest
This work was supported by the China Postdoctoral Science Foundation Funded Project (2015M570407), the National Basic Research Program of China (973 Program) (2012CB725202), the National Natural Science Foundation of China (21276110, 31500065), the High-tech Research and Development Programs of China SS2015AA021004, 2014AA021304), the Research Project of Chinese Ministry of Education (113033A), the Fundamental Research Funds for the Central Universities (JUSRP51306A, JUSRP11545), the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, the 111 Project (111-2-06) and the Jiangsu province “Collaborative Innovation Center for Advanced Industrial Fermentation” industry development program.
REFERENCES
|
Abrunhosa L, Venâncio A (2007). Isolation and purification of an enzyme hydrolyzing ochratoxin A from Aspergillus niger. Biotechnol. Lett. 29(12):1909-1914.
Crossref
|
|
|
|
Austin BP, József T, Péter B (2011). The substrate specificity of Metarhizium anisopliae and Bos taurus carboxypeptidases A: Insights into their use as tools for the removal of affinity tags. Protein Expr Purif. 77:53-61.
Crossref
|
|
|
|
|
Bhutto MA (2011). Optimization of cultural conditions for protease production by Bacillus subtilis EFRL 01. Afr. J. Biotechnol. 10: 5173-5181
|
|
|
|
|
Cheng TC, Vijay R, Chan SI (1999). Purification and characterization of a cobalt-activated carboxypeptidase from the hyperthermophilic archaeon Pyrococcus furiosus. Prot. Sci. 8:2474-2486.
Crossref
|
|
|
|
|
Bown DP, Gatehouse JA (2004). Characterization of a digestive carboxypeptidase from the insect pest corn earworm (Helicoverpa armigera) with novel specificity towards C-terminal glutamate residues. Eur. J. Biochem. 271:2000-2011
Crossref
|
|
|
|
|
Drzymala A, Bielawski W (2009). Isolation and characterization of carboxypeptidase III from germinating triticale grains. Acta Biochim Biophys Sin. 4:69-78.
Crossref
|
|
|
|
|
Elena K, Reeta B, Iryna B (2007). A novel subfamily of mouse cytosolic carboxypeptidases. FASEB J. 21: 836-850.
Crossref
|
|
|
|
|
Fang L, Yasuda M (2005). Debittering effect of Monascus carboxypeptidase during the hydrolysis of soybean protein, J. India. Microbiol. Biotechnol. 32:487-489.
Crossref
|
|
|
|
|
Gao X, Cui W, Tian Y (2013). Over-expression, secretion, biochemical characterization, and structure analysis of Bacillus subtilis aminopeptidase. J. Sci. Food Agric. 93:2810-2815.
Crossref
|
|
|
|
|
Gao X, Liu Z, Cui W (2014). Enhanced thermal stability and hydrolytic ability of Bacillus subtilis aminopeptidase by removing the thermal sensitive domain in the non-catalytic region. PLoS One 9(3):e92357.
Crossref
|
|
|
|
|
Garten W, Klenk HD (1983). Characterization of the carboxypeptidase involved in the proteolytic cleavage of the influenza haemagglutinin. J. Gen. Virol. 64:2127-2137.
Crossref
|
|
|
|
|
Imtiaz S (2013). Production of alkaline protease by Bacillus subtilis using solid state fermentation. Afr. J. Microbiol. 7:1558-1568.
|
|
|
|
|
Jia M, Xu M, He B (2013). Cloning, expression, and characterization of L-asparaginase from a newly isolated Bacillus subtilis B11-06. J. Agric. Food Chem. 61:9428-9434.
Crossref
|
|
|
|
|
Joshi L, Leger RJS (1999). Cloning, expression, and substrate specificity of MeCPA, a zinc carboxypeptidase that is secreted into infected tissues by the fungal entomopathogen Metarhizium anisopliae. J. Biol. Chem. 274:9803-9811.
Crossref
|
|
|
|
|
Kazuhisa H, Kamisoyama H (2000). Purification and characterization of Carboxypeptidase A from chicken pancreas. J. Anim. Sci. 71:520-523.
|
|
|
|
|
Kumar A, Sharma A, Sharma R (2014). Identification, characterization and analysis of expression of gene encoding carboxypeptidase A in Anopheles culicifacies A (Diptera: culicidae). Acta Trop. 139:123-130.
Crossref
|
|
|
|
|
Lee HS, Kim YJ, Bae SS, Jeon JH, Lim JK, Kang SG, Lee J-H (2006). Overexpression and characterization of a Carboxypeptidase from the hyperthermophilic Archaeon Thermococcus sp. NA1. Biosci. Biotechnol. Biochem. 70(5):1140-1147.
Crossref
|
|
|
|
|
Li X, Solomon B (1997). Thermal stabilization of Carboxypeptidase A as a function of pH and ionic milieu. Biochem. Mol. Biol. Int. 43:60l-611.
Crossref
|
|
|
|
|
Lyons P, Callaway ML (2008). Characterization of carboxypeptidase A6, an extracellular matrix peptidase. J. Biol. Chem. 283: 7054-7063.
Crossref
|
|
|
|
|
Schallmey M, Singh A, Ward OP (2004). Developments in the use of Bacillus species for industrial production. Can. J. Microbiol. 50:1-17.
Crossref
|
|
|
|
|
Nagamori Y, Kusaka K, Fujishima N (1991). Enzymatic properties of dipeptidyl carboxypeptidase from Bacillus pumilus. Agric. Biol. Chem. 55:1695-1699.
Crossref
|
|
|
|
|
Pereira PJ, Segura-Martın S, Oliva B, Ferrer-Orta C, Aviles FX, Coll M, Gomis-Rüth FX, Vendrell J (2002). Human Procarboxypeptidase B: Three-dimensional Structure and Implications for Thrombin-activatable Fibrinolysis Inhibitor (TAFI). J. Mol. Biol. 321:537-547.
Crossref
|
|
|
|
|
Sebastian T, Julia L, Javier GP, Degroeve S, Lennart M, Francesc XA, Gevaert K, Petra VD (2013). Proteome-derived Peptide Libraries to Study the Substrate Specificity Profiles of Carboxypeptidases. Mol. Cell Proteom. 12:096-2110
|
|
|
|
|
Suwen W, Sonia S, Josep V (2002). Identification and characterization of three members of the human metallocarboxypeptidase gene family. J. Biol. Chem. 277:14954-14964.
|
|
|
|
|
Tardioli PW, Fernándezâ€Lafuente R, Guisan JM (2003). Design of new immobilized-stabilized carboxypeptidase A derivative for production of aromatic free hydrolysates of proteins. Biotechnol. Prog. 19:565-574.
Crossref
|
|
|
|
|
Vertesi A, Kiss I S B, Simon L M (1999). Preparation, characterization and application of immobilized carboxypeptidase A. Enzyme Microb. Technol . 25:73-79.
Crossref
|
|
|
|
|
Vijayalakshmi K, Rajakumar S, Vijayalakshmi K (2011). Antimicrobial protein production by Bacillus amyloliquefaciens MBL27: Optimization of culture conditions using Taguchi's experimental design. Indian J. Sci. Technol. 4:0974-6846.
|
|
|
|
|
Yoshinaka R, Sato M, Morishita J (1985). Enzymic Characterization of Carboxypeptidase A from the Catfish Pancreas. Bull. Jap. Soc. Sci. Fish. 51:113-116.
Crossref
|
|