ABSTRACT
In 2015, a disease of unknown origin appeared in Torreon, Coahuila, in the Northeast of Mexico, causing great mortality in Phoenix palms, especially in Phoenix canariensis. Until early 2019, around 1300 palms died from this disease. The aim of this study was to determine its etiology. The symptoms registered in affected palms were similar to those described for Texas Phoenix Palm Decline (TPPD). Phytoplasmas were detected in samples from nine P. canariensis individuals using a TaqMan/real-time PCR assay specific for group 16SrIV detection. DNA of positive samples was amplified by nested PCR using primer pair P1/P7 followed by LY16Sf/LY16-23Sr and R16F2n/R16R2. In silico analysis of the sequences obtained revealed the presence of phytoplasmas associated with TPPD, belonging to subgroup 16SrIV-D. This is the first report of a disease associated with subgroup 16SrIV-D phytoplasmas in the Northeast of Mexico, further extending the known geographical range of this pathogen.
Key words: Texas Phoenix palm decline, 16SrIV-D phytoplasmas, nested polymerase chain reaction (PCR), sequence analysis.
Lethal yellowing-type diseases (LYDs) associated with group 16SrIV phytoplasmas are among the most important diseases affecting palms worldwide due to the significant economic losses that they have caused throughout the years, particularly in coconut (Cocos nucifera L.) plantations (Gurr et al., 2016). Since no successful method for culturing group 16SrIV phytoplasmas on artificial media exist, these pathogens are currently studied using a variety of detection techniques that target phytoplasma DNA in infected host tissues, such as nested PCR (Gundersen and Lee, 1996), which is useful for phytoplasma characterization and in silico sequence analysis, and real-time PCR (Córdova et al., 2014), which increases sensitivity and reduces sample-processing time.
In addition to C. nucifera, LYDs are known to affect several other palm species including edible date (Phoenix dactylifera L.) and Canary Island date (Phoenix canariensis hort. ex Chabaud) (Harrison and Elliott, 2016). These two species of palms have been used as ornamentals for more than sixty years in the city of Torreon, Coahuila, Mexico. In 2015, some of these plants began showing symptoms suggestive of a phytoplasma disease, similar to both lethal yellowing (LY) and one of its variants, the Texas Phoenix palm decline (TPPD) (McCoy et al., 1980; Harrison et al., 2008). By early 2019, the disease in Torreon was associated with the death of around 1300 Phoenix species palms throughout the city, mostly affecting mature P. canariensis individuals.
The phytoplasmas associated with LY and TPPD, subgroups 16SrIV-A and D, respectively, are the most commonly reported group 16SrIV strains affecting palm and non-palm species in the United States (Harrison et al., 2008; Bahder et al., 2019), Mexico (Oropeza et al., 2020), Central America and the Caribbean region (Ntushelo et al., 2013; Myrie et al., 2014). Although P. canariensis and P. dactylifera can be affected by both subgroups (Harrison et al., 2008), observations made in Florida suggest that they are more susceptible to subgroup 16SrIV-D (Harrison and Elliott, 2016). Also, both phytoplasma subgroups have been detected in natural populations of one of their candidate vectors, the planthopper Haplaxius crudus Van Duzee (Narváez et al., 2018), whose occurrence in the urban area of Torreon has been reported recently (Hernández-Rodríguez et al., 2019). However, despite the fact that mortality in P. canariensis in association with subgroup 16SrIV-A and D phytoplasmas has been reported on numerous occasions in Southern United States (Harrison et al., 2002c, 2008; Ong and McBride, 2009; Singh, 2014), so far, there are no reports in scientific journals of either of these phytoplasmas affecting P. canariensis palms in Mexico, nor has an outbreak of LY or TPPD occurred in the country’s entire Northeast region (comprising the states of Coahuila, Nuevo Leon and Tamaulipas). Therefore, the objective of this study was to determine, by means of nested PCR and real-time PCR detection, followed by in silico sequence analysis, if the disease affecting Phoenix palms in Torreon, Coahuila, is associated with a phytoplasma enclosed in group 16SrIV.
Sampling and evaluation of palms
Between December 2015 and October 2018, trunk samples were collected from eleven Canary Island date palms with symptoms suggesting a LYD at different locations in the city of Torreon, state of Coahuila (25°32′22″N, 103°26′55″W). P. canariensis was selected for our study as it became evident that most symptomatic palms in Torreon belonged to this species. All sampled individuals as well as other P. canariensis showing identical symptoms were monitored closely in order to record and describe symptom progression. Samples were obtained using a portable electric drill following Oropeza et al. (2010). After collection, samples were stored on an ice box for transportation to the laboratory, where they were refrigerated (usually 1 to 3 days) until nucleic acid extraction. Samples from five asymptomatic P. canariensis were also included in this study for comparative purposes.
DNA extraction
Total DNA was extracted by modifying the previously described method of Doyle and Doyle (1990). 1 g of trunk tissue ground to a powder with liquid nitrogen was incubated in 5 mL of CTAB extraction buffer (2% CTAB/100 mM Tris-HCl pH 8.0/20 mM EDTA pH 8.0/1.4 M NaCl/1% PVP) added with 0.1% 1-Thioglycerol, for 30 min at 65°C. A volume of chloroform-isoamyl alcohol (24:1) was added and the mix was stirred. To precipitate the supernatant obtained after centrifugation, isopropanol and sodium acetate (3 M) were used, followed by incubation at -20°C for 1 h. Lastly, the DNA pellet was re-suspended in 30 to 50 µL of TE buffer (10 mM Tris/1 mM EDTA).
Phytoplasma detection by real-time PCR
Detection analyses for group 16SrIV phytoplasmas were performed by a TaqMan/real-time PCR assay following the protocol described by Córdova et al. (2014). Reactions were carried out using a Rotor-Gene® Q thermocycler (QIAGEN). The following amplification parameters were used: an initial phase of 2 min at 50°C followed by 10 min at 95°C, then 40 cycles of amplification, each with a first denaturation step at 95°C for 15 s and a second alignment step at 61°C for 1 min. The Ct (cycle threshold) value of each sample was assigned by manually adjusting the threshold line to intersect with the exponential phase of the amplification curves and automatically setting the baseline using the Rotor-Gene Q® - Pure Detection software version 2.0.2 (QIAGEN). A Ct cut-off value of 32 was used for discriminating positive and negative samples in accordance with Narváez et al. (2017). All samples were assessed in duplicate. Experimental controls consisted of DNA from a LY-infected Pritchardia pacifica Seem. and H. Wendl. palm (positive) and ultrapure water (negative).
Phytoplasma detection by nested PCR
Samples that were positive for the presence of phytoplasma DNA by the TaqMan/real-time PCR assay were subjected to nested PCR in order to obtain amplicons for sequencing purposes. For the first amplification, phytoplasma universal primers P1/P7 (Deng and Hiruki, 1991; Schneider et al., 1995) were used, followed by a second round of amplification with universal primers R16F2n/R16R2 (Lee et al., 1993; Gundersen and Lee, 1996) or the group specific set LY16Sf/LY16-23Sr (Harrison et al., 2002c, b). All reactions were performed in a T100™ thermocycler (Bio-Rad). The amplification programs for the reactions with the P1/P7 and R16F2n/R16R2 primer pairs were carried out following the specifications of Cordova-Lara et al. (2017), while the reactions with the LY16Sf/LY16-23Sr pair were performed according to Harrison et al. (2002a). The P1/P7 products were diluted 1:20 or 1:40 with ultrapure water prior to the second amplification, for both sets of primers. Positive and negative controls were included in all reactions as described previously.
Cloning and sequencing
Amplification products were purified with the QIAquick® Gel Extraction Kit (QIAGEN) and cloned into the pGEM®-T Easy Vector System I (Promega) following the manufacturers’ instructions. To purify the recombinant plasmids, the QIAprep® Spin Miniprep Kit (QIAGEN) was used. Cloned inserts were sent to the University of California, Davis (The College of Biological Sciences UCDNA Sequencing Facility) for subsequent sequencing.
In silico analysis
The F2nR2 fragment of all sequences was analyzed using the iPhyClassifier interactive online tool (Zhao et al., 2009). Virtual RFLP profiles were generated and similarity coefficients were calculated to classify the phytoplasma strains detected based on previously established criteria (Wei et al., 2008). Subsequent molecular analyses were performed with MEGA software version X (Kumar et al., 2018). A phylogenetic tree was constructed with representative phytoplasma 16S rRNA gene sequences applying the maximum likelihood method based on the General Time-Reversible model for nucleotide substitution. The reliability of the analysis was subjected to a bootstrap test with 1000 replicates.
Symptom description
P. canariensis palms with symptoms suggestive of a LYD (Figure 1) were monitored in Torreon, Coahuila. The observed progression of symptoms was as follows: in healthy or asymptomatic fruit-bearing palms usually all leaves were green (except those going through natural senescence) and inflorescences were bearing fruit (Figure 1A). At the onset of symptoms, infected palms started showing discoloration of the oldest leaves; yellowing was observed first, quickly followed by bronzing and desiccation (Figure 1B). This decay process continued to the middle canopy leaves (Figure 1C). During this period of foliar decay there was also premature fruit drop (Figure 1D), with inflorescences becoming fruitless and dry (Figure 1E), and in some cases, death of the spear leaf (Figure 1F). Finally, leaf deterioration reached the upper leaves (Figure 1G) and ended with the decay of the entire foliage (Figure 1H) and collapse of the crown (Figure 1I). In some plants, roots were revised, and decay was also observed (not shown).
Phytoplasma detection in date palms
Nine out of eleven symptomatic P. canariensis palms that were sampled tested positive to the group 16SrIV phytoplasma detection analysis with the TaqMan/real-time PCR assay; sampling dates and Ct values obtained for each palm are shown in Table 1. Five asymptomatic palms were also analyzed and no amplification was obtained. The DNA from the nine positive P. canariensis was then subjected to nested PCR to obtain amplicons for subsequent sequencing and phytoplasma identification. Positive amplifications were obtained for two samples with primer pair LY16Sf/LY16-23Sr and for three others with primer pair R16F2n/R16R2; no amplification was obtained for the rest of the samples.
Analysis of phytoplasma rDNA
Five phytoplasma sequences from P. canariensis were obtained and deposited in the GenBank® database of the National Institutes of Health (NIH). Primers R16F2n/R16R2 produced three 1249 bp sequences (Accession numbers MN384667, MN384668 and MN384670) and LY16Sf/LY16-23Sr produced two 1745 bp sequences (Accession numbers MN389510 and MN607700). BLAST® comparison of these sequences showed that the highest sequence identity matches were with sequences of subgroup 16SrIV-D phytoplasmas (Table 2).
Phytoplasma identification at the subgroup level was confirmed by analyzing in detail the F2nR2 fragment of the sequences obtained with iPhyClassifier. All sequences from Coahuila generated an identical virtual RFLP profile. This pattern was most similar to the one generated by the reference sequence of subgroup 16SrIV-D (Accession number AF237615), with a similarity coefficient of 0.98. Closer examination of the virtual gels revealed that the only difference between both patterns is produced by the BfaI enzyme (Figure 2). Based on the criteria established by Wei et al. (2008), the phytoplasmas detected in the P. canariensis palms that were included in this study all belong to subgroup 16SrIV-D.
A phylogenetic analysis was carried out with phytoplasma 16S rRNA gene sequences (Figure 3). All Coahuila sequences were grouped in the same clade, which in turn was grouped into a larger one shared with other members of the 16SrIV-D and B subgroups as well. When examining the alignment used for the construction of the phylogenetic tree, a single base substitution was found at position 1286 of the 16S rRNA gene (in the reference sequence of subgroup 16SrIV-D, Acc. AF237615) that distinguishes the Coahuila strains from the rest of the subgroup 16SrIV-D phytoplasma sequences that were included in the matrix (Figure 4).
In the Americas, LYDs affecting C. nucifera and several other palms, including important ornamental species like P. canariensis, have been associated with group 16SrIV phytoplasmas (Ntushelo et al., 2013). In 2015, a disease with symptoms suggestive of a LYD appeared in Torreon, Coahuila, causing great mortality in Phoenix palms, especially in P. canariensis individuals around fifty to sixty years old. In this study, detection protocols based on nested PCR and real-time PCR were used to investigate the presence of group 16SrIV phytoplasmas in symptomatic P. canariensis individuals sampled in Torreon during 2015 to 2018. Our results showed that this disease is associated with phytoplasmas belonging to subgroup 16SrIV-D, thus representing a new outbreak of TPPD rather than LY. This diagnosis is consistent with the symptoms observed in affected palms (Figure 1) according to the descriptions given by Harrison et al. (2002c) and Harrison and Elliott (2016). This is the first report of a disease associated with subgroup 16SrIV-D phytoplasmas in the Northeast of Mexico. Also, P. canariensis is reported for the first time as a host of this subgroup outside of the United States.
The majority of the P. canariensis sampled in our study (82%) were positive to the group 16SrIV detection analyses according to real-time PCR results. To classify these phytoplasma strains, five nested PCR products obtained with primer pairs LY16Sf/LY16-23Sr and R16F2n/R16R2, were cloned and sequenced. BLAST® analysis of these sequences suggested an identity with subgroup 16SrIV-D phytoplasmas. All sequences produced an identical virtual RFLP profile when analyzed with iPhyClassifier; this pattern was more similar to that of the reference sequence of subgroup 16SrIV-D (similarity coefficient of 0.98), with the only difference between them exposed by enzyme BfaI in the virtual digestion. This change is likely the result of a single base substitution located at position T/C1286 of the 16S rRNA gene (Figure 4), since it provides an extra C^TAG recognition site for the enzyme and thus shortens the largest fragment produced in the virtual digestion (Figure 2). Information gathered so far suggest this substitution could be used to further differentiate between subgroup 16SrIV-D strains present in different regions, but more exploration is needed to support this hypothesis. In addition, a phylogenetic tree was constructed with phytoplasma 16S rRNA gene sequences. Coahuila sequences were grouped in the same clade, which, in turn, grouped into a larger one with other members of the 16SrIV-D and B subgroups present in other regions. This shows the genetic proximity of the phytoplasmas detected in Coahuila, which although they do not constitute a new subgroup within group 16SrIV, they can be clearly distinguished from other members of the 16SrIV-D subgroup in a phylogenetic tree based on differences in the virtual RFLP profile and the presence of point mutations in the sequence corresponding to the F2nR2 fragment of the 16S rRNA gene, a similar case to the geographical variants of subgroup 16SrIV-B strains reported affecting C. nucifera palms in the Dominican Republic (Martinez et al., 2008).
P. canariensis was the first species to show TPPD-like symptoms in Torreon, also the most affected during the outbreak, therefore our focus on this palm, however, later inspection of P. dactylifera individuals also present in the city and displaying similar symptoms (data not shown) suggest that TPPD is affecting this species as well, a prospect that needs confirmation. As of early 2019, this disease has been associated with the death of around 1300 Phoenix spp. palms in several parts of the city, where approximately 2000 Phoenix palms were estimated to be present before the onset of the disease (Samaniego-Gaxiola et al., unpublished data). To our knowledge, this is also the first reported outbreak of a LYD associated with subgroup 16SrIV-D phytoplasmas in Mexico with an unusually high apparent mortality rate.
Subgroup 16SrIV-D was first detected in the mid-1990s affecting Carludovica palmata Ruiz and Pav. plants at Calkini, Campeche (Cordova et al., 2000). In the late 1990s, it was detected again infecting coconuts palms on the Pacific coast of Mexico (Harrison et al., 2002b). This was followed by subsequent reports in a number of regions including South Texas (Harrison et al., 2002c), West Central Florida (Harrison et al., 2008) and the Ticul-Merida area in Yucatan (Vázquez-Euán et al., 2011). This pathogen is now widely distributed throughout Mexico and Southern United States according to available literature (Figure 5), however, it remains difficult to explain its presence in places as distant as the Miami-Dade County in Florida (Bahder et al., 2019) and the Bay of La Paz in Baja California Sur (Poghosyan et al., 2019), considering its absence in intermediate locations like the majority of Mexico’s northern states.
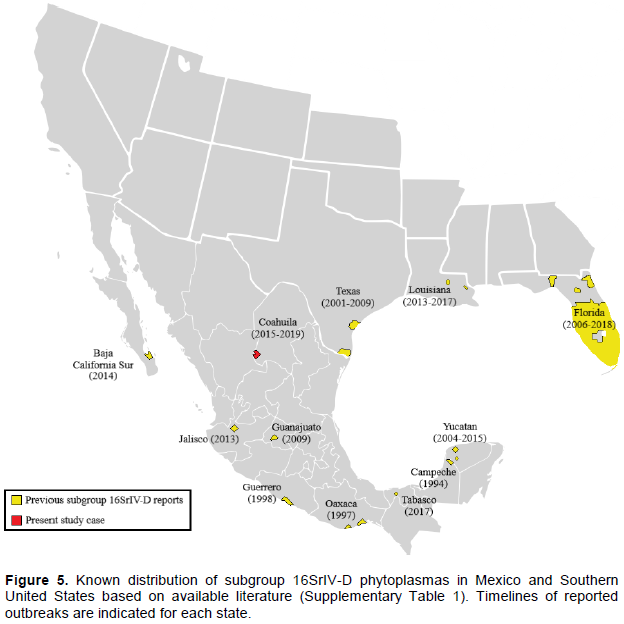
The origin of the Torreon outbreak is difficult to infer, however, due to the ample presence of P. canariensis in the city for more than sixty years, it seems unlikely that subgroup 16SrIV-D phytoplasmas have been established in the area for a long time, given the high susceptibility of this host to TPPD (Harrison and Elliott, 2016). The pathogen could have arrived with a recent introduction of contaminated plant material (possibly ornamental palms), or through natural spread by a vector. In 2009, a syndrome with similar characteristics was observed in P. dactylifera and Sabal mexicana Mart. palms located in the state of Guanajuato, Mexico (Aviña-Padilla et al., 2011). It should be noted that the phytoplasma sequences obtained by the authors of that study (Accession Numbers JF431249 and JF431250) also have a single base substitution located at position T/C1286 of the 16S rRNA gene, unfortunately, the length of the Guanajuato sequences is significantly shorter than those of Coahuila, so they could not be included in the phylogenetic analysis conducted in this study to establish a possible relationship between them. It remains to be seen whether these two cases of TPPD are connected in some way, or if the disease reached Torreon from somewhere else, given also its relative proximity to the Cameron, Hidalgo and Willacy counties in Texas, where TPPD is considered active (Ong and McBride, 2009).
Since 2015, declining Phoenix palms in Torreon have been removed or treated with antibiotics in an effort to mitigate the impact of the disease. These measures continue to be relevant in light of the recent discovery of a putative vector of TPPD, H. crudus, in the urban area of Torreon (Hernández-Rodríguez et al., 2019). This implies that the disease could affect other susceptible palms in the city, like the already mentioned P. dactylifera as well as Syagrus romanzoffiana (Cham.) Glassman, which has been reported on multiple occasions as a host of subgroup 16SrIV-D in Florida (Harrison et al., 2008; Bahder et al., 2018, 2019). In the meantime, ornamental species with similar characteristics to date palms but no prior history of susceptibility to TPPD can now be suggested for reforestation purposes, such as Washingtonia filifera (Linden ex André) H. Wendl. and Washingtonia robusta H. Wendl.
The finding of subgroup 16SrIV-D in Coahuila, as well as recently in Louisiana (Singh and Ferguson, 2017), Baja California Sur (Poghosyan et al., 2019) and Tabasco (Ramos et al., 2020), greatly extends the known geographical range of this pathogen (Figure 5), indicating an increased spread of TPPD in recent years. This situation requires the immediate attention of competent authorities and the scientific community in the United States and Mexico in view of the potential spread of TPPD to major date production states like California (USA) or Sonora (Mexico) (Wright, 2016; Ortiz-Uribe et al., 2019) or to other cities within those countries that use Phoenix palms as ornamentals.
The authors have not declared any conflict of interests.
This work was carried out in the frame of European Union’s Horizon 2020 research and innovation programme project “Tropicsafe” under grant agreement number 727459. The authors thank CONACYT (Mexico) for the scholarship no. 466618 awarded to P. J. Palma-Cancino and Ana M. Castruita for her help during sample collection.
REFERENCES
|
Aviña-Padilla K, Rodríguez-Páez LA, Nava-Castrejón ÁI, Ochoa-Sánchez JC, Rivera-Bustamante R, Martínez-Soriano JP (2011). Epidemic of lethal yellowing disease affecting Phoenix dactilyfera and Sabal mexicana in central Mexico. Bulletin of Insectology 64(Supplement):S221-S222.
|
|
|
|
Bahder BW, Helmick EE, Chakrabarti S, Osorio S, Soto N, Chouvenc T, Harrison NA (2018). Disease progression of a lethal decline caused by the 16SrIV-D phytoplasma in Florida palms. Plant Pathology 67(8):1821-1828.
Crossref
|
|
|
|
|
Bahder BW, Soto N, Helmick EE, Dey KK, Komondy L, Humphries AR, Mou DF, Bailey R, Ascunce MS, Goss EM (2019). A survey of declining palms (Arecaceae) with 16SrIV-D phytoplasma to evaluate the distribution and host range in Florida. Plant Disease 103(10):2512-2519.
Crossref
|
|
|
|
|
Cordova I, Oropeza C, Almeyda H, Harrison NA (2000). First report of a phytoplasma-associated leaf yellowing syndrome of palma jipi plants in southern México. Plant Disease 84(7):807.
Crossref
|
|
|
|
|
Cordova-Lara I, Mota-Narváez L, Puch-Hau C, Oropeza C, Sáenz L (2017). Detection and identification of lethal yellowing phytoplasma 16SrIV-A and D associated with Adonidia merrillii palms in Mexico. Australasian Plant Pathology 46(5):389-396.
Crossref
|
|
|
|
|
Córdova I, Oropeza C, Puch-Hau C, Harrison N, Collí-Rodríguez A, Narvaez M, Nic-Matos G, Reyes C, Sáenz L (2014). A real-time PCR assay for detection of coconut lethal yellowing phytoplasmas of group 16srIV subgroups A, D and E found in the Americas. Journal of Plant Pathology 96(2):343-352.
|
|
|
|
|
Deng S, Hiruki C (1991). Amplification of 16S rRNA genes from culturable and nonculturable Mollicutes. Journal of Microbiological Methods 14(1):53-61.
Crossref
|
|
|
|
|
Dey KK, Jeyaprakash A, Hansen J, Jones D, Smith T, Davison D, Srivastava P, Bahder B, Li C, Sun X (2018). First report of the 16SrIV-D phytoplasma associated with decline of a Bismarck palm (Bismarckia nobilis). Plant Health Progress 19(2):128.
Crossref
|
|
|
|
|
Doyle JJ, Doyle JL (1990). Isolation of plant DNA from fresh tissues. Focus 12(1):13-15.
|
|
|
|
|
González-Pacheco BE, Rojas-Martínez RI, Ochoa-Martínez DL, Silva-Rosales L (2014). First report of a 16Sr IV group phytoplasma associated with lethal yellowing in Agave tequilana. Journal of Plant Pathology 96(3):603.
|
|
|
|
|
Gundersen DE, Lee I-M (1996). Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathologia Mediterranea 35(3):144-151.
|
|
|
|
|
Gurr GM, Johnson AC, Ash GJ, Wilson BAL, Ero MM, Pilotti CA, Dewhurst CF, You MS (2016). Coconut lethal yellowing diseases: a phytoplasma threat to palms of global economic and social significance. Frontiers in Plant Science 7:1521.
Crossref
|
|
|
|
|
Harrison NA, Elliott ML (2016). Phytoplasmas associated with date palm in the continental USA: three 16SrIV subgroups. Emirates Journal of Food and Agriculture 28(1):17-23.
Crossref
|
|
|
|
|
Harrison NA, Helmick EE, Elliott ML (2008). Lethal yellowing-type diseases of palms associated with phytoplasmas newly identified in Florida, USA. Annals of Applied Biology 153(1):85-94.
Crossref
|
|
|
|
|
Harrison NA, Helmick EE, Elliott ML (2009). First report of a phytoplasma-associated lethal decline of Sabal palmetto in Florida, USA. Plant Pathology 58(4):792.
Crossref
|
|
|
|
|
Harrison NA, Myrie W, Jones P, Carpio ML, Castillo M, Doyle MM, Oropeza C (2002a). 16S rRNA interoperon sequence heterogeneity distinguishes strain populations of palm lethal yellowing phytoplasma in the Caribbean region. Annals of Applied Biology 141(2):183-193.
Crossref
|
|
|
|
|
Harrison NA, Narvaez M, Almeyda H, Cordova I, Carpio ML, Oropeza C (2002b). First report of group 16SrIV phytoplasmas infecting coconut palms with leaf yellowing symptoms on the Pacific coast of Mexico. Plant Pathology 51(6):808.
Crossref
|
|
|
|
|
Harrison NA, Womack M, Carpio ML (2002c). Detection and characterization of a lethal yellowing (16SrIV) group phytoplasma in Canary Island date palms affected by lethal decline in Texas. Plant Disease 86(6):676-681.
Crossref
|
|
|
|
|
Hernández-Rodríguez S, Valdés-Perezgasga MT, López-Hernández J, García F, Hernández V, Obrador-Sánchez JA (2019). Cixiidos (Hemiptera: Cixiidae) asociados a palmas con síntomas del amarillamiento letal del cocotero (ALC) en el área urbana de Torreón, Coahuila, México. Entomología Mexicana 6:526-529.
|
|
|
|
|
Jeyaprakash A, Sutton BD, Halbert SE, Schubert TS (2011). First report of a 16SrIV-D phytoplasma associated with Texas Phoenix palm decline on pigmy date palm (Phoenix roebelenii) in Florida. Plant Disease 95(11):1475.
Crossref
|
|
|
|
|
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018). MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35(6):1547-1549.
Crossref
|
|
|
|
|
Lee I-M, Hammond RW, Davis RE, Gundersen DE (1993). Universal amplification and analysis of pathogen 16S rDNA for classification and identification of mycoplasmalike organisms. Phytopathology 83(8):834-842.
Crossref
|
|
|
|
|
Martinez RT, Narvaez M, Fabre S, Harrison NA, Oropeza C, Dollet M, Hichez E (2008). Coconut lethal yellowing on the southern coast of the Dominican Republic is associated with a new 16SrIV group phytoplasma. Plant Pathology 57(2):366.
Crossref
|
|
|
|
|
McCoy RE, Miller ME, Thomas DL, Amador J (1980). Lethal decline of Phoenix palms in Texas associated with mycoplasmalike organisms. Plant Disease 64(11):1038-1040.
Crossref
|
|
|
|
|
Myrie WA, Harrison NA, Douglas L, Helmick EE, Gore-Francis J, Oropeza C, McLaughlin WA (2014). First report of lethal yellowing disease associated with subgroup 16SrIV-A phytoplasmas in Antigua, West Indies. New Disease Reports 29:12.
Crossref
|
|
|
|
|
Narváez M, Ortíz E, Silverio C, Santamaría JM, Espadas F, Oropeza C (2017). Changes observed in Pritchardia pacifica palms affected by a lethal yellowing-type disease in Mexico. African Journal of Biotechnology 16(51):2331-2340.
|
|
|
|
|
Narváez M, Vázquez-Euán R, Harrison NA, Nic-Matos G, Julia JF, Dzido JL, Fabre S, Dollet M, Oropeza C (2018). Presence of 16SrIV phytoplasmas of subgroups A, D and E in planthopper Haplaxius crudus Van Duzee insects in Yucatán, Mexico. 3 Biotech 8:61.
Crossref
|
|
|
|
|
Ntushelo K, Harrison NA, Elliott ML (2013). Palm phytoplasmas in the Caribbean Basin. Palms 57(2):93-100.
|
|
|
|
|
Ong K, McBride S (2009). Palm diseases caused by phytoplasmas in Texas. NPDN Second National Meeting December 6-10, 2009 Miami, Florida.
|
|
|
|
|
Oropeza C, Sáenz L, Narvaez M, Nic-Matos G, Córdova I, Myrie W, Ortiz CF, Ramos E (2020). Dealing with lethal yellowing and related diseases in coconut. In: Coconut Biotechnology: Towards the Sustainability of the 'Tree of Life', S Adkins, M Foale, R Bourdeix, Q Nguyen, J Biddle (Eds.). Switzerland; Springer Nature.
Crossref
|
|
|
|
|
Oropeza C, Narváez M, Echegoyén-Ramos PE, Rodas R (2010). Plan de Contingencia Ante un Brote de Amarillamiento Letal del Cocotero (ALC) en un País de la Región del OIRSA. Organismo Internacional Regional de Sanidad Agropecuaria - OIRSA. San Salvador, El Salvador, 149 p.
|
|
|
|
|
Ortiz-Uribe N, Salomón-Torres R, Krueger R (2019). Date palm status and perspective in Mexico. Agriculture 9(3):46.
Crossref
|
|
|
|
|
Poghosyan A, Hernandez-Gonzalez J, Lebsky V, Oropeza C, Narvaez M, Leon de la Luz JL (2019). First report of 16SrIV palm lethal yellowing group phytoplasma ('Candidatus Phytoplasma palmae') in palmilla de taco (Brahea brandegeei) and palma colorada (Washingtonia robusta) in the state of Baja California Sur, Mexico. Plant Disease 103(8):2122.
Crossref
|
|
|
|
|
Ramos E, Lesher-Gordillo JM, Oropeza C, Ortiz-García CF, Magaña-Alejandro MA, Sánchez-Soto S, García-Estrada Y (2020). Detection and identification of phytoplasmas in the 16SrIV-A, -B and -D subgroups in palms in Tabasco, Mexico. Plant Disease (In press).
Crossref
|
|
|
|
|
Schneider B, Seemueller E, Smart CD, Kirkpatrick BC (1995). Phylogenetic classification of plant pathogenic mycoplasma-like organisms or phytoplasmas. In: Molecular and Diagnostic Procedures in Mycoplasmology, Vol. I., Razin S, Tully JG (Eds.). San Diego, CA; Academic Press pp:369-380.
Crossref
|
|
|
|
|
Singh R (2014). Texas Phoenix palm decline confirmed in Louisiana. NPDN News 9(1):1-2.
|
|
|
|
|
Singh R, Ferguson MH (2017). First report of a 'Candidatus Phytoplasma palmae'-related subgroup 16SrIV-D phytoplasma on Trachycarpus fortunei. Australasian Plant Disease Notes 12:59.
Crossref
|
|
|
|
|
Vázquez-Euán R, Harrison NA, Narvaez M, Oropeza C (2011). Occurrence of a 16SrIV group phytoplasma not previously associated with palm species in Yucatan, Mexico. Plant Disease 95(3):256-262.
Crossref
|
|
|
|
|
Wei W, Lee I-M, Davis RE, Suo X, Zhao Y (2008). Automated RFLP pattern comparison and similarity coefficient calculation for rapid delineation of new and distinct phytoplasma 16Sr subgroup lineages. International Journal of Systematic and Evolutionary Microbiology 58(10):2368-2377.
Crossref
|
|
|
|
|
Wright GC (2016). The commercial date industry in the United States and Mexico. HortScience 51(11):1333-1338.
Crossref
|
|
|
|
|
Zhao Y, Wei W, Lee I-M, Shao J, Suo X, Davis RE (2009). Construction of an interactive online phytoplasma classification tool iPhyClassifier, and its application in analysis of the peach X-disease phytoplasma group (16SrIII). International Journal of Systematic and Evolutionary Microbiology 59(10):2582-2593.
Crossref
|
|