ABSTRACT
Molecular analysis in plants requires the use of high quality DNA. Currently, there are a myriad of DNA extraction protocols, which still seems that their applicability across different species is not straightforward. The aim of the present study was to evaluate four different cetyl trimethylammonium bromide (CTAB)-based DNA extraction protocols for Heliconia species from lyophilized fully mature leaves. A split-plot design with two levels (six different species and four DNA extraction methods) was used, where treatments were distributed in a randomized block design with four replications. The response variables were total yield (µg) and purity (A260/A280 nm), and were determined by UV-spectrophotometry and agarose gel electrophoresis, respectively. Additionally, EcoRI enzyme restriction digestion and amplification of the Hc_D6 locus marker were used to evaluate the functionality of the extracted DNA. Analysis of variance (ANOVA) results indicated that the extraction methods had a significant effect on the DNA yield (F = 8.51, df = 3, P<0.0001) and purity (F = 10.43, df = 3, P<0.0001). The best methods to obtain DNA from Heliconia spp. were those described by Michiels et al. (2003) and Sagahi-Maroof et al. (1984) with modifications.
Key words: DNA extraction, Heliconia, tropical plants, cetyl trimethylammonium bromide (CTAB), polyvinylpyrrolidone (PVP).
Current protocols in molecular biology require the use of high quality DNA. For some plant species, this rather common process can turn into a burdensome task, due to the presence of secondary metabolites. These compounds may undergo rapid oxidation, binding tightly to the DNA and subsequently, co-precipitate with the DNA preventing its use in downstream applications (Porebski et al., 1997; Sheperd et al., 2002; Shabnam and Saeed, 2016; Swati et al., 2016). Tropical plants for example, often have high levels of polysaccharides, polyphenols, proteins, lipids, among others, which make the isolation and purification of DNA difficult (Barra et al., 2012; Cavallari et al., 2014; Huang et al., 2014). In addition, the presence of these compounds in the isolated DNA inhibits the activity of various enzymes used in molecular biology experiments, such as DNA ligases, polymerases and endonucleases, affecting their efficiency, and in many cases rendering negative results (Friar, 2005; Cavallari et al., 2014; Swati et al., 2016).
The research team of this study worked on the population genetics of the native heliconia species from the southern part of Mexico (Avendaño-Arrazate, 2017). Heliconias are tropical plants that have been cultivated as ornamentals outside their natural distribution areas, increasing their importance as cut flowers. The genus Heliconia L. displays a great array of diversity of species, varieties, hybrids and cultivars, often leading to taxonomic inaccuracies among scientists and commercial growers (Marouelli et al., 2010). The majority of the studies have focused mostly on taxonomy (Iles et al., 2017), regeneration and in vitro propagation and very few on morphological (Arrazate-Avendaño et al., 2017) and molecular characterization (Marouelli et al., 2010; Isaza et al., 2012). Therefore, more molecular studies are needed to clarify these discrepancies and the isolation of high quality DNA is one of the central steps to achieve it.
Although there are few reports about extracting DNA from these species, in some cases it requires the use of robotics (Côrtes et al., 2009), additional purification steps after the DNA extraction, using commercial kits (Meléndez-Ackerman et al., 2005) or the use of phenol-chloroform cleaning steps (Kumar et al., 1998; Sheela et al., 2006). Therefore, the objective of this study was to compare four commonly used cetyl trimethylammonium bromide (CTAB) DNA extraction methods with modifications: Haque et al. (2008) Method 1; Saghai-Maroof et al. (1998) Method 2; Doyle and Doyle (1990) Method 3 and Michiels et al. (2005) Method 4. The starting material was mature leaves and the effectiveness of the methods was tested based on yield, purity, integrity and functionality, in order to determine the most suitable method for isolating high yield of quality DNA from Heliconia spp.
Plant
Fully developed leaves were collected from the “Rosario Izapa Experimental Field Station” (Chiapas, Mexico). Four different heliconia species were used: Heliconia stricta Hubert, Heliconia wagneriana Peterson, Heliconia bourgaeana and Heliconia collinsiana Griggs. Additionally, leaf samples from Canna indica L. species belonging to the Zingeberales order, were also included as control. After harvest, leaf tissue was kept at -80°C for at least 48 h prior to lyophilization. Freeze-dried tissue was ground into fine powder using the TissueLyser II (Qiagen, Hilden, Germany).
Experimental design
A split-plot design with two levels (six different species and four DNA extraction methods) was used to determine the best extraction method for heliconias. The big plots were the DNA extraction methods, and the different species were the split-plots. Treatments were distributed in a randomized block design with four replications. Three samples per species in each method were extracted (12 samples of the same species per replicate). In total, 240 DNA extractions were carried out. The response variables were total yield (µg) and purity (A260/A280 nm). All DNA extractions were carried out following the same procedure to control the variation inherent to the different steps of each method. Statistical analyses were performed using SAS PROC GLM to test the effect of the extraction method on DNA yield and purity. Means were compared using the Tukey test with a significance level of 0.05 (SAS, 2011).
DNA extraction
All DNA extractions were carried out with 50 mg of lyophilized tissue in a 2.0 ml tube. Extraction buffer (1 ml) was added to each tube followed by incubation at 65°C for 45 min with constant mixing (Table 1). Tubes were allowed to cool at room temperature (RT) for about 5 min and to each tube, 700 µl of chloroform:isoamyl alcohol (24:1) were added. The tubes were inverted gently for 10 min to form an emulsion and were centrifuged at 10,000 × g for 10 min at RT. The upper phase was decanted into a new tube and the washing step was repeated. After the second centrifugation, about 600 µl were transferred to a new 2.0 ml tube containing 5 µl of RNaseA (10 mg/mL) and incubated at 37°C for 30 min. DNA was precipitated with 600 µl of cold isopropanol, mixing gently by inversion several times until DNA was visible.
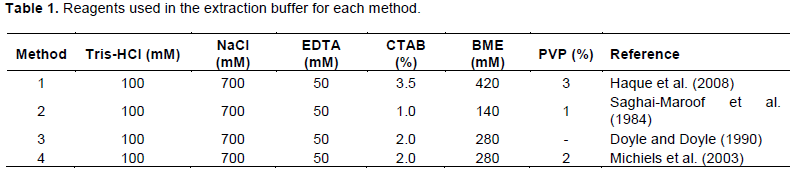
The tubes were centrifuged at 10,000 × g for 10 min at RT and the supernatant was carefully poured off. The DNA pellets were washed twice with ethanol at 70 and 95%, for 5 min and were spun again as before. The DNA pellets were allowed to dry at RT and were resuspended in 300 µl of TE buffer (10 mM Tris-HCl pH 8.0; 1 mM EDTA pH 8.0) overnight.
Measurement of DNA quantity, purity and integrity
DNA concentration was measured using the NanoDrop 2000 UV-Vis Spectophtometer (Thermo Scientific, Wilmington, DE, USA). The A260/A280 nm absorbance ratio was used to assess the purity of the nucleic acid samples (Held, 2001). Integrity of the extracted DNA was evaluated by electrophoresis of 100 ng/well for each sample on 1% agarose gel. Migration distance was compared to that of 100 ng of uncut lambda DNA (NEB, Ipswich, MA, USA).
Restriction endonuclease digestion
To assess the functionality of the extracted DNA for downstream applications, 50 ng of DNA were digested with 15 units of EcoRI (NEB, Ipswich, MA, USA) in a 20 µl reaction for 5 h at 37°C. Digested DNA was assessed by visual inspection on a UV transluminator (Kodak Gel Logic 112, Woodbridge, CT, USA) after 1% agarose gel electrophoresis and GelRed™ staining (Biotium, Hayward, CA, USA).
PCR amplification
DNA amplification reactions of the H. caribaea specific microsatellite marker (Hc_D6) were performed only for Heliconia spp. samples (C. indica was excluded since this marker is specific for Heliconia) in a 10 µl reaction (Gowda et al., 2012). The PCR cocktail consisted of 10 ng of DNA, 4 µl of 2X RedTaq® Ready Mix™ (SIGMA-ALDRICH, St. Louis, MO, USA), 0.5 µM of each primer (For 5′ACT GCA CAT CAT ATC ATC CTG3′ and Rev 5′GTG GGT CAG TCA ATT ACT GTG3′) and sterilized water. The expected size of the Hc_D6 product was 222 to 258 bp. Amplifications were carried out on a PrimeG Thermal Cycler (Techne Inc., Burlington, NJ, USA). The amplification was carried out with an initial denaturation step of 94°C for 5 min, followed by 30 cycles of 30 s at 94°C, 30 s at 58°C, 30 s at 72°C and a final extension step of 6min at 72°C. PCR amplification products were visualized as described previously.
DNA yield
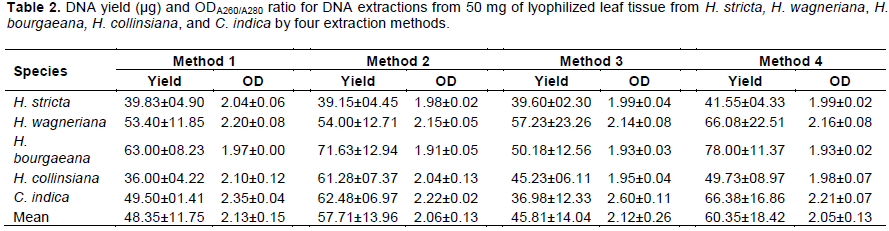
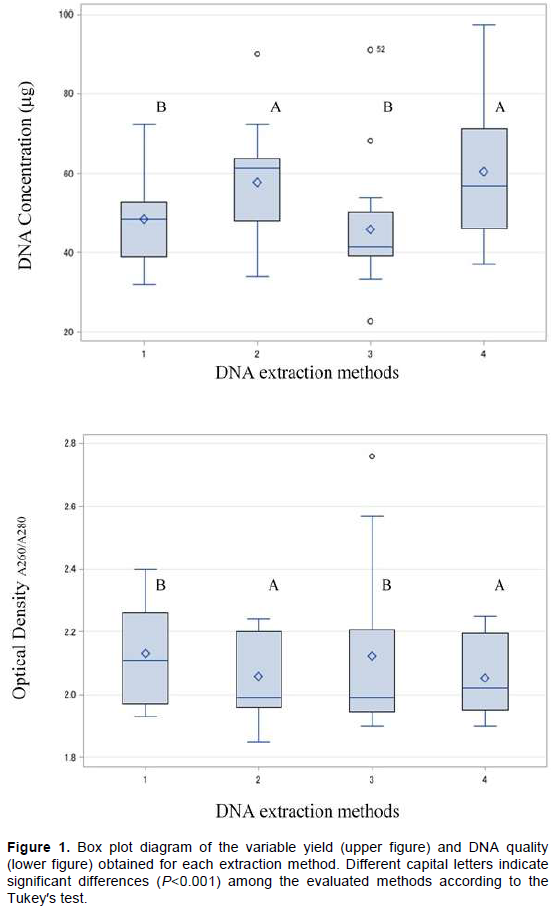
Yield of the extracted DNA by the four different methods is shown in Table 2. The extraction methods had a significant effect on the DNA yield (F = 8.51, df = 3, P<0.0001). In general, the extraction methods that contain polyvinylpyrrolidone (PVP, MW 40,000, Sigma-Aldrich, St. Louis, MO, USA), had significantly higher DNA yields. Methods 4 and 2 (Michiels et al., 2003; Saghai-Maroof et al., 1984) were statistically superior for extracting DNA in the evaluated species. Each of these methods yielded on average 60.35±18.42 and 57.70±13.96 µg, compared to Method 3, with 45.84±14.04 µg, which had the lowest yield (Figure 1). Methods 2 and 4 included PVP in their extraction buffer while Method 3 did not. PVP has been reported to form insoluble complexes with lactones and phenolic compounds (Kim et al., 1997; Barra et al., 2012), and the presence of these compounds in Heliconia spp. have been detected in different studies (Estrada et al., 2008; Hernández-Meneses et al., 2013). Additionally, through the centrifugation steps after each the addition of chloroform: isoamyl alcohol, these compounds are removed by precipitation (Haque et al., 2008; Swati et al., 2016; Abdel-Latif and Osman, 2017). These steps, in combination with the use of CTAB, aided the selective precipitation of DNA, which could explain the higher concentrations of DNA obtained by these methods.
DNA purity and integrity
The purity of the DNA obtained was evaluated as a function of the ODA260/A280 ratio (Held, 2001). The mean ODA260/A280 ratios for the four methods evaluated in this research ranged from 2.05 to 2.13 (Table 2). The statistical analysis indicates that the extraction methods had a significant effect on the DNA purity (F = 10.43, df = 3, P<0.0001), where Methods 2 and 4 had the narrowest range for the ODA260/A280 ratio (Figure 1). Method 3 had the highest ODA260/A280 ratio values (2.76, outlier value in Figure 1); however, all these values came from the C. indica samples (which had a similar trend regardless of the method). In general, Methods 1, 2 and 4 had ODA260/A280 ratio values within the commonly accepted range (1.8 to 2.0). These values can be explained by the removal of polysaccharides and proteins, usually bound to the DNA. With the addition of NaCl in the extraction buffer, the solubility of these compounds is increased and the use of the chloroform: isoamyl alcohol wash steps reduces their co-precipitation with the DNA molecule. Furthermore, the inclusion of the RNaseA step in all methods, helps to decrease the absorbance of these compounds during the spectrophotometric readings (Tamari et al., 2013; Abdel-Latif and Osman, 2017).
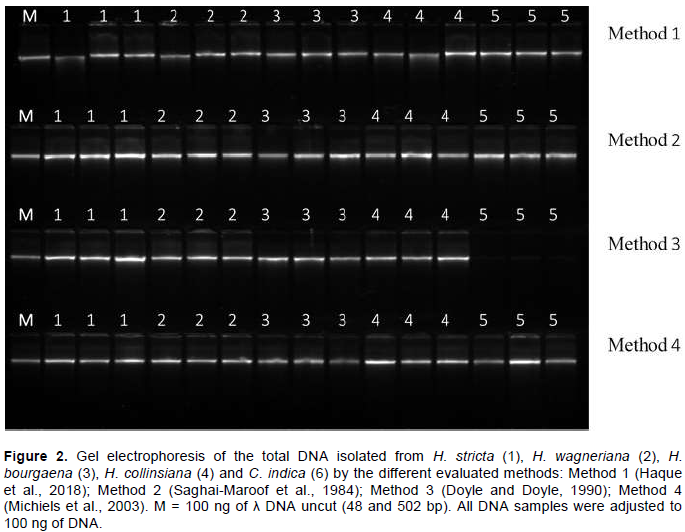
Since the ODA260/A280 ratio is only an indication of DNA purity rather than a precise answer, the integrity (taken as DNA of high molecular weight) of the extracted DNA samples was determined by electrophoresis on a 1% agarose gel. Figure 2 shows that all protocols were equally efficient for obtaining high DNA quality and integrity in Heliconia spp. (from the qualitative point of view). However, the analysis of variance showed that for C. indica, Method 3 was not as efficient as the others. In fact, the DNA yield for this method was the lowest as compared to the other methods (Table 2) and had the highest mean ODA260/A280 ratio (2.60±0.11). One possible explanation for this result might be the presence of high levels of phenolic compounds in C. indica (Vankar and Srivasta, 2008), and the use of the extraction buffer of Method 3. This buffer did not include PVP and was not as effective as the other buffers for the removal of phenolic compounds. The presence of these compounds in the DNA samples can interfere with the quantification by UV absorption, causing problems during the estimation of the concentrations of the DNA samples and shifting the ODA260/A280 ratio (Mannin, 1991).
DNA functionality
One of the most important features of any DNA extraction protocol is whether the extracted DNA will work in downstream applications. Conventionally, this feature is evaluated digesting the extracted DNA (Barra et al., 2012; Huang et al., 2013; Cavallari et al., 2014). Figure 3 shows the electrophoresis pattern of the DNA samples fully digested with EcoRI restriction enzyme. There is an absence of high molecular weight bands, and the continuous characteristic smearing of the different fragment sizes is present. It indicated that the DNA obtained by all the evaluated methods is of high purity and quality. Amplification of the Hc_D6 locus marker from the total DNA extracted by each method produced a clear and clean target product of 222 to 258 bp in length (Figure 4). This further confirmed the quality of the DNA, free of any inhibitory compounds and suitable for any downstream application (sequencing reactions were also performed successfully, data not shown). At first glance, the DNA extraction protocols described here seem equally efficient in extracting high yield and quality of genomic DNA from the five analyzed species. Yet, once statistical tools were applied into the analysis of results, significant differences were found for both variables. Methods 4 (Michiels et al., 2003) and Method 2 (Saghai-Maroof et al., 1984) with the modifications assayed in this study, were significantly different for extracting higher DNA yields and better DNA quality. Unlike other protocols, all the steps described here can be carried out at RT, eliminating the need for refrigerated centrifugation or DNA precipitation. Additionally, all the DNA samples were found to be stable when stored at -20°C for more than two years and could be amplified by PCR.
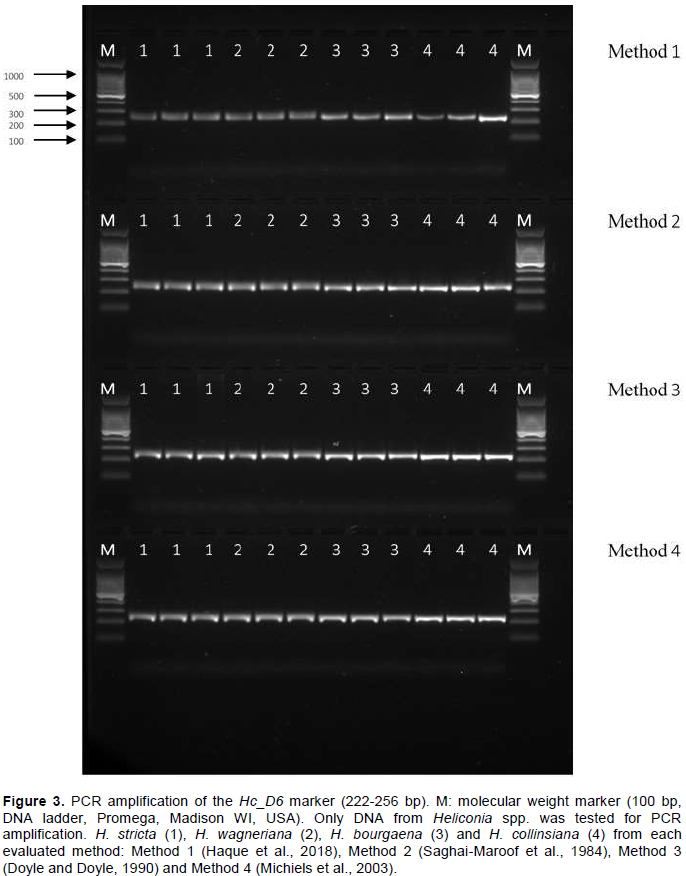
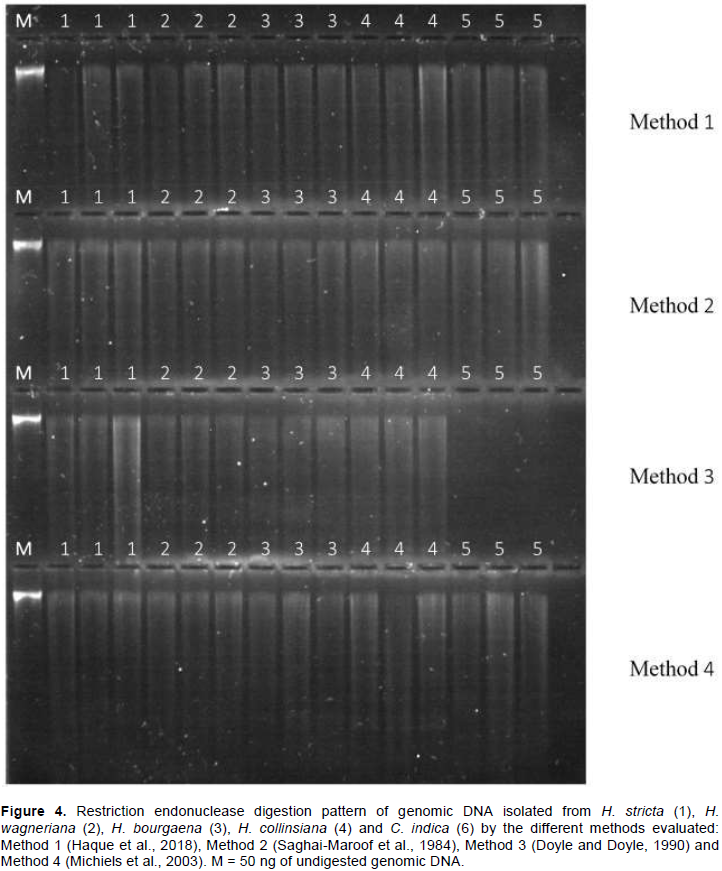
Although all evaluated protocols were efficient for DNA extraction from mature leaves of Heliconia spp., Methods 4 (Michiels et al., 2003) and 2 (Saghai-Maroof et al., 1984) were statistically superior for obtaining high yield and quality DNA. These methods yielded reproducible amplification of SSR products, demonstrating its compatibility for molecular analysis. These protocols are routinely used in our laboratory to isolate DNA from a wide variety of plant species including Amaranthus hypocondriacus L., Amaranthus cruentus L., Agave species, Theobroma cacao L., Stevia rebaudiana Bert, Pinus species, Pseudotsuga menziesii (Mirb) Franco, and Annona muricata L., all having high levels of secondary metabolites.
The authors have not declared any conflict of interests.
This research was supported by the National Forestry, Crops and Livestock Research Institute of Mexico, through the project “Rescue and Conservation of Heliconias from Southern Mexico”.
REFERENCES
|
Abdel-Latif A, Osman G (2017). Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize. Plant Methods 13:1-9.
Crossref
|
|
|
|
Avenda-o-Arrazate CH, Arrazate-Argueta, JA, Ortiz-Curiel S, Moreno-Pérez E, Iracheta-Donjuan L, Reyes-López D, Grajales-Solís M, Martínez-Bola-os M, Cortés-Cruz M (2017). Morphological characterization in wild species of Heliconias (Heliconia spp) in Mexico. American Journal of Plant Sciences 6:1210-1223.
|
|
|
|
|
Barra M, Salzar E, Beltrán M Sagredo B (2012). Simple and robust extraction method for the large-scales analysis of genotypes containing high polyphenolic content, such as landraces of Solanum tuberosum and Zea mays. Ciencia e investigación agraria 39:593-601.
Crossref
|
|
|
|
|
Cavallari MM, Siqueira MVBM, Val TM, Pavanelli JC, Monteiro M, Granado C, Pinheiro JB, Zucchi MI, Gimenez MA (2014). A modified acidic approach for DNA extraction from plant species containing high levels of secondary metabolites. Genetics and Molecular Research13:6497-6502.
Crossref
|
|
|
|
|
Côrtes MC, Gowda V, Kress WJ, Bruna EM, Uriarte M (2009). Characterization of 10 microsatellite markers for the understory Amazonian herb Heliconia acuminate. Molecular Ecology Resources 1261-1264.
Crossref
|
|
|
|
|
Doyle JJ, Doyle JL (1990). Isolation of plant DNA from fresh tissue. Focus 12:13-15.
|
|
|
|
|
Estrada GS, Quintana JC, Jiménez SL, Alarcón PJC, Perea-ez JJA, Vargas LJ (2009). Evaluación fitoquímica preliminar de Heliconia psittacorum y Heliconia rostrata y de la potencial actividad inhibitoria de algunos de los efectos del veneno de Bothropsasper (mapaná X). Vitae 2:252-257.
|
|
|
|
|
Friar EA (2005). Isolation of DNA from plants with large amounts of secondary metabolites in enzymology. Methods in Enzymology 395:32-38.
Crossref
|
|
|
|
|
Gowda V, Erickson DL, Kress WJ (2012). Development and characterization of microsatellite loci for two Caribbean Heliconia (Heliconiaceae: H. bihai and H. caribaea). American Journal of Botany 99(2):e81-e83.
Crossref
|
|
|
|
|
Haque IR, Bandopadhyay R, Mukhopadhyay K (2008). An optimized protocol for fast genomics DNA isolation from high secondary metabolites and gum containing plants. Asian Journal of Plant Sciences 7:304-308.
Crossref
|
|
|
|
|
Held P (2001). Nucleic acid purity assessment using A260/A280 ratios. Application Note.
View . Accessed on Dec 5, 2017.
|
|
|
|
|
Hernández-Meneses E, López-Peralta MCG, Estrada-Luna AA (2013). Callogénesis de Heliconia collinsiana Griggs in vitro: establecimiento, inducción y proliferación. Revista Mexicana de Ciencias Agrícolas 8:1175-1186.
|
|
|
|
|
Huang QX, Wang XC, Kong H, Guo Y-L, Guo AP (2014). An efficient DNA isolation method for tropical plants. African Journal of Biotechnology12(19):2727-2732.
|
|
|
|
|
Iles WJD, Sass CH, Lagomarsino L, Benson-Martin G, Driscoll H, Specht CD (2017). The phylogeny of Heliconia (Heliconiaceae) and the evolution of floral presentation. Molecular Phylogenetics and Evolution.117:150-167.
Crossref
|
|
|
|
|
Isaza L, Marulanda ML, López AM (2012). Genetic diversity and molecular characterization of several Heliconia species in Colombia. Genetics and Molecular Research 11:4552-4563.
Crossref
|
|
|
|
|
Kim CS, Lee CH, Shin JS, Chung YS, Hyung NI (1997). A simple and rapid method of isolation of high quality genomic DNA from fruit trees and conifers using PVP. Nucleic Acids Research 25(5):1085-1086.
Crossref
|
|
|
|
|
Kumar PP, Yau JCK, Goh CJ (1998). Genetic analysis of Heliconia species and cultivars with randomly amplified polymorphic DNA (RAPD) markers. Journal of the American Society for Horticultural Science 123(1):91-97.
|
|
|
|
|
Marouelli LP, Inglis PW, Ferreira MA, Buso GSC (2010). Genetic relationships among Heliconia (Heliconiaceae) species based on RAPD markers. Genetics and Molecular Research 9(3):1377-1387.
Crossref
|
|
|
|
|
Mannin L (1991). Isolation of nucleic acids from plants by different solvent precipitation. Analytical Biochemistry 195(1):45-50.
Crossref
|
|
|
|
|
Meléndez-Ackerman EJ, Speranza P, Kress WJ, Rohena L, Toledo E, Cortés C, Treece D, Gitzendanner M, Soltis P, Soltis D (2005). Microevolutionary processes inferred from AFLP and morphological variation in Heliconia bihai (Heliconiaceae). International Journal of Plant Sciences 166(5):781-794.
Crossref
|
|
|
|
|
Michiels AE, Van den Ende W, Tucker M, Van Riet L, Van Laere A (2003). Extraction of high-quality genomic DNA from latex-containing plants. Analytical Biochemistry 315(1):85-89.
Crossref
|
|
|
|
|
Porebski S, Bailey L, Baum B (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and phenol components. Plant Molecular Biology Reporter 15(1):8-15.
Crossref
|
|
|
|
|
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984). Ribosomal DNA spacer-length polymorphism in barley: Mendelian inheritance, chromosomal location and population dynamics. Proceedings of the National Academy of Sciences of the United States of America 81(24):8014-8019.
Crossref
|
|
|
|
|
SAS Institute Inc. (2011). Base SAS 9.3 Procedures Guide. Cary NC: SAS Institute Inc.
|
|
|
|
|
Shabnam A, Saeed A (2016). An efficient and simple CTAB method for total genomic DNA isolation from low amounts of aquatic plants with a high level of secondary metabolites. Progress in Biological Sciences 6(1):95-106.
|
|
|
|
|
Sheela VL, Geetha Lekshmi PR, Jayachandran Nair CS, Rajmohan K (2006). Molecular characterization of Heliconia by RAPD assay. Journal of Tropical Agriculture 44(1-2):37-41.
|
|
|
|
|
Sheperd M, Cross M, Stokoe RL, Scott LJ, Jones ME (2002). High-throughput DNA extraction from forest trees. Plant Molecular Biology Reporter 20(4):425-425.
Crossref
|
|
|
|
|
Swati R, Geeta JD, Annapurna AN, Arunkumar N, Karaba N (2016). Standardization of DNA extraction method from mature dried leaves and ISSR-PCR conditions for Melia dubia Cav. A fast growing multipurpose tree species. American Journal of Plant Sciences 7(3):437-445.
Crossref
|
|
|
|
|
Tamari F, Hinkley CS, Ramprashad N (2013). A comparison of DNA extraction methods using Petunia hybrid tissues. Journal of Biomolecular Techniques 24(3):113-118.
|
|
|
|
|
Vankar PS, Srivastava J (2008). Comparative study of total phenol, flavonoid contents and antioxidant activity in Canna indica an Hibiscus rosasinensis: Prospective natural food dyes. International Journal of Food Engineering 4(3):1-16.
Crossref
|
|