ABSTRACT
Burkina Faso livestock is made up of two main cattle population, namely Zebuine and Taurine. Transhumance and settlement of Zebu cattle breeders in tsetse challenged areas lead to cross-breeding Zebu and Taurine. Introgression of the Zebu cattle may have changed the structure of the trypanotolerant Lobi/Baoulé breed. The objective of the present study was to appreciate the introgression of Zebu genes into Baoulé population by assessing the structure and the genetic diversity of cattle populations across the tsetse belt in Burkina Faso. Therefore, 450 blood samples were taken for genotyping in 29 villages of 3 main regions where Baoulé, Baoulé×Zebu and Zebu populations are found. Twenty five loci of 22 autosomes have been genotyped. The mean of observed alleles per locus was 12.44±4.31 while the mean of expected alleles was 4.67±1.48. The heterozygosity ranged from 0.34 to 0.76 and 0.36 to 0.87, respectively for observed and expected heterozygosity across loci. The average heterozygosity across population was 0.73±0.10. The mean estimates of F-statistics were FIS = 0.117±0.019, FIT = 0.158±0.019 and FST = 0.047±0.005. The phylogenetic tree showed the Baoulé South-West segregating apart from the other populations, Baoulé×Zebu being an intermediate genetic group between Baoulé South-West and Zebu North populations. The Baoulé West could not be differentiated from crosses. The Baoulé breed seems to be impacted by the introgression of Zebu genes to its biotope and pure Baoulé seems to be confined to the South-West with very few pure individuals in the West.
Key words: Burkina Faso, Zebu, Baoulé×Zebu, Baoulé, introgression, microsatellite.
African cattle populations are said to be originated from 2 wild aurochs populations (Loftus et al., 1994, 1999; Bradley et al., 1994). Bos taurus (taurine), the humpless descendants of aurochs were domesticated in either the Near East or on the African continent (Epstein, 1971; Clutton-Brock, 1989; Bradley et al., 1996; Hanotte et al., 2002). Several investigations indicated that African Zebu cattle are an admixture of Bos indicus and B. taurus (MacHugh et al., 1997; Hanotte et al., 2002). Analysis of mitochondrial DNA sequences and microsatellites loci indicate that B. indicus may have diverged from B. taurus (Bradley et al., 1996; MacHugh et al., 1997; Hanotte et al., 2002). In West Africa, cattle populations are representative of both shorthorn (B. taurus brachyceros) and longhorn (B. taurus longifrons) humpless taurines, humped zebus (B. indicus) and Zebu/Taurine cattle (Gautier et al., 2009).
In Burkina Faso, indigenous cattle are very important for the subsistence and economic development of the country. These indigenous cattle provide essential food products, draft power, manure, and income for rural people. Indigenous breeds are well adapted to local environment thus they have developed disease tolerance and adaptation to harsh climatic conditions. This adaptation favoured the survival under stresses and exploitation of poor quality feeds stuff (Sodhi et al., 2005; Gautier et al., 2009).
With different drought episodes in 1973 and 1983 that occurred in Burkina Faso (Paturel et al., 1998) and the shift of the Northern limit lines of tsetse flies (Courtin et al., 2010) there has been an introgression of Zebu cattle genes through the movement of pastoralist people in the tsetse challenged areas (Grace et al., 2007) seeking for grass and water for livestock. In addition, some of the transhumant livestock keepers settled for long in the tsetse challenged areas rearing and crossing the Zebu breed to the local taurine to control the recurrent trypanosomosis disease. Local mixed livestock-crops farmers crossbreed also the Zebu to the local taurine since the 1920s to 1930s (Grace, 2005) to get hybrid animals that are used as draught animals. The intermediate sized animal is preferred because the local taurine is smaller and less powerful. These trends may have changed the structure of the populations in the tsetse challenged zones. It was therefore important to ascertain the introgression of Zebu genes into Baoulé breed in order to help guide decisions on improvement and conservation priorities. This is especially necessary owning to the husbandry systems practiced by local livestock farmers, which may affect diversity levels through high gene flow between breeds.
Animals
1045 blood samples were taken from animals belonging to Baoulé also named Lobi cattle, Zebu and crossbred of Baoulé×Zebu cattle populations (471 males and 574 females) out of which 450 samples have been randomly selected as per location for genotyping. The animals have been sampled in 29 villages of three different regions and different altitudes. The North (5 villages) being a tsetse free region, the South-West and the West are tsetse challenged regions (Figure 1). Tsetse free is in upper part of the map and separated from the tsetse challenged regions by the northern limit lines of Glossina tachinoides, Glossina morsitans submorsitans and Glossina palpalis gambiensis
Six populations have been considered in the analysis (Table 1); Zebu of the challenged areas have been merged (Other Zebu) due to low number (4) of Zebu samples in the West. Baoulé×Zebu population in the West was the biggest sample out of the six populations. That results from crossbreeding the 2 main breed (Zebu and Baoulé) to control trypanosomosis disease in the tsetse challenged areas of Burkina Faso in general. Crossbreed population had the highest size in the genotyped sampled (158). On the other hand, the trypanotolerant Baoulé was more important in terms of size in the South-West than the other regions.
DNA extraction
Whole blood of each individual was dropped onto a Whatman FTA card according to Whatman protocol BD09. The samples were kept in multi-barrier pouch till punching day.
Three millilitres diameter Harris punch has been used to remove sample discs from the spotted cards. Genomic DNA was isolated according to a modified protocol of Whatman (Soudré, 2011).
DNA amplification
Microsatellites (31) primers were chosen for the amplification of the genomic DNA. 15 were donated by the International Livestock Research Institute, Nairobi, Kenya. PCR conditions were optimized and all the 31 microsatellites tested for polymorphism. A final panel of 25 polymorphic microsatellites has been used for genotyping of the cattle populations (BM1818, BM1824, BM2113, CSSM066, ETH3, ETH10, ETH185, ETH225, HAUT24, HAUT27, HEL1, HEL5, HEL9, HEL13, ILSTSS005, ILSTS006, INRA023, INRA032, TGLA53, TGLA122, TGLA126, TGLA227, AGLA293, ILSST033 and MGTG4B). Microsatellites were selected combining information from both the National Centre for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/) database and BOVMAP (http://locus.jouy.inra.fr/cgi-bin/bovmap/intro2.pl) covering 22 autosomal chromosomes regions. PCRs were then performed using Applied Biosystems GeneAmp® PCR System 9700 thermal cycler.
Genotyping process
The PCR products were diluted 1/10 in distilled water, and genotyping was performed on MegaBACETM 500, fluorescence-based DNA system utilizing capillary electrophoresis. Alleles were called and scored under MegaBACETM Genetic Profiler Software Suite v2.2 system.
Statistical analysis
Estimates of total number of alleles, mean number of alleles, effective number of alleles, observed heterozygosity (Ho), and unbiased gene diversity (expected unbiased heterozygosity, He) for each population were obtained with POPGENE program version 1.31 (Yeh et al., 1999). He the most common measure of variability (Petit et al., 1998; Caballero and Toro, 2002) was estimated using the algorithm of Levene (1949), which is the same as Nei’s (1987) unbiased heterozygosity. Convert package version 1.31 (Glaubitz, 2004) has been used to determine the allele frequencies and detect breed-specific alleles.
Deviations from Hardy-Weinberg Equilibrium (HWE) probability exact test with unbiased exact P-value of Guo and Thompson (1992) was performed using GENEPOP package version 4.0.10 of Rousset (2008) according to the Markov Chain parameters, dememorization (1000), batches (100), and iteration per batch (1000).
Using the variance-based of Weir and Cockerhan (1984), F-statistics (FIS, FIT, FST) for each locus and overall values were calculated using FSTAT version 2.9.3.2 (Goudet, 2002). Significance tests on the estimates F-statistics for each microsatellite locus were obtained by constructing 95 and 99% confidence intervals based on the standard deviations estimated by jackknifing across populations using FSTAT.
POPULATION 1.2.30 software (Langella, 1999) was used to construct a phylogenetic tree of populations with bootstrap on locus using Reynolds et al. (1983) least squares. That was run using UPGMA and 1000 trials. The tree was visualized with TreeView 1.6.6 software (Page, 1996).
The Bayesian clustering method, as implemented by the STRUCTURE 2.3.3 program (Pritchard et al., 2000; Falush et al., 2003, 2007; Hubisz et al., 2009) was run five times with burnin period of 5.104 iterations followed by 105 number of MCMC repeats after burnin assuming k=2. The admixture model was used with the sampling locations as a prior. Correlated allele frequencies model was used as well. The clustering has also been performed with Bayesian Analysis of Population Structure (BAPS) package version 5.3 (Corander et al., 2008) that showed the same pattern as STRUCTURE.
Alleles (311) were detected from the 25 loci surveyed, giving a mean number of alleles 12.44 ± 4.31 observed and mean of effective alleles number was 4.67 ± 1.48 (Table 1). The number of alleles ranged from 3 at CSSM066 to 22 at TGLA122 (Table 2). The lowest number of effective allele number was observed at CSSM066 (1.57) and the highest number at TGLA53 (7.65). The total number of alleles per population ranged from 176 to 270 alleles, respectively for Baoulé in the West and Zebu Baoulé in the West. The lowest number of alleles in Baoulé West population may be due to the small size of the Baoulé population (21) from this region in the sample. The total number of allele in taurine was somehow higher than in Zebuine may be because most of the primers are taurine based designed. Many have been designed from Hereford for example.
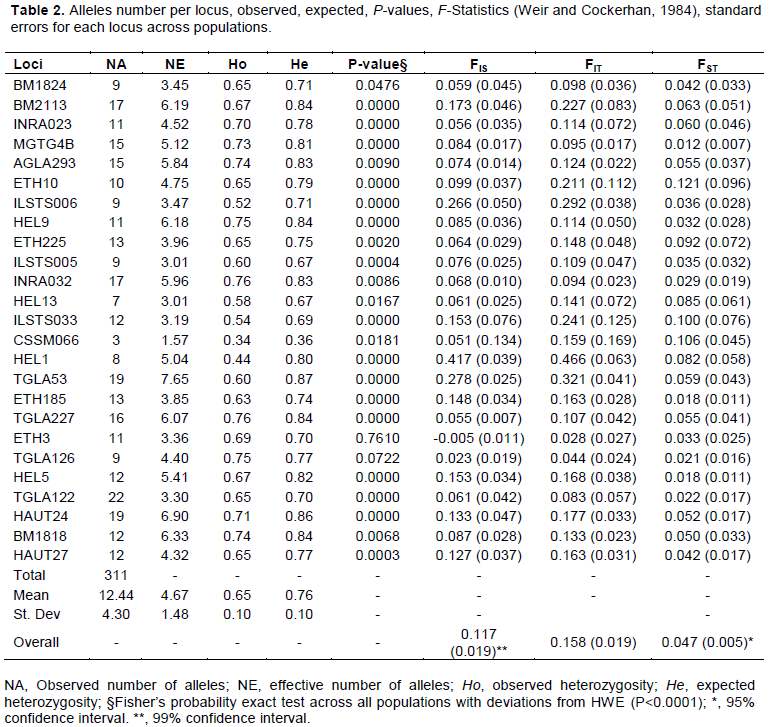
Some 7% of the total alleles have been detected as breed-specific alleles using Convert package, 12 private alleles of Zebu at 11 loci and 10 private alleles of Baoulé at 9 loci.
For loci in the study, 14 of them have deviated from HWE (P<0.0001 using the probability exact test of Fisher) as shown in Table 2; markers displaying a highly significant deviation from HWE are in italics. Furthermore, deviations from HWE were statistically significant (P<0.0001) for 26 locus-population combinations. Out of the 25 loci analyzed in each population, 1 to 8 deviated significantly from HWE. All populations also deviated from HWE (P<0.0001) probably because of heterozygote’s deficiency.
Expected heterozygosity has been generally higher than observed heterozygosity not only at marker level but also at population level. At population level, the most diversified population was the Zebu Baoulé in the West with the highest observed and expected heterozygosity (Ho = 0.68±0.10, He = 0.77±0.08) and the less diversified the Baoulé from the South-West (Ho = 0.60±0.14, He = 0.68±0.14). In the overall population, the crosses as expected are more diversified (He = 0.77±0.09) than the pure breeds (Zebu and Baoulé) followed by Zebu (He = 0.74±0.11). Observed heterozygosity ranged from 0.34 (CSSM066) to 0.76 (INRA032 and TGLA227). The most variable marker in this study was TGLA53 (He = 0.87) compared to CSSM066 (He = 0.36). The marker which contributed much to the variability was INRA032 and HEL 9 (Ave. Het. = 0.82). The mean number of migrants per generation for all loci estimated based on the formula Nm = 0.25(1-FST)/FST as implemented in POPGENE was 6.06.
Baoulé×Zebu population from the South-West showed the highest value of FIS (0.157). Comparisons of FIS of the 3 groups were not statistically significant as well as the comparisons of the FST values.
The locus ETH10 (0.121) contributed the most to the population differentiation. But the overall FST being < 0.15, the population differentiation seems to be moderate. The mean global FST ranged from 0.012 (MGTG4B) to 0.121 (ETH10) among different microsatellite loci with an estimated mean value of 0.047 (P<0.01), indicating 4.7% of the total variation being attributed to between breed differences.
A neighbor-joining dendogram constructed based on unbiased genetic distances showed 2 main clusters, one cluster composed of Baoulé South-West and the second being composed of the remaining populations (Figure 2). In the second cluster, the populations clustered further into 3 genetic groups; the first group had Baoulé×Zebu South-West and Other Zebu. That group had the smallest genetic distance (DA = 0.0434 in Table 3). The last group had only Zebu North. The unbiased genetic distance between Baoulé South-West and Zebu North was the longest one (DA = 0.3390). A phylogenetic tree (Figure 2) supports it with a bootstrap value of 75%. The bootstraps showed the Baoulé South-West segregating from the other populations with 100% of replicates.
Using STRUCTURE, the most likely K is that where ln Pr(G⁄K) is maximized. The maximum value of ln Pr(G⁄K) was obtained at K = 2 (Figure 3), that provided an explanation of the genetic structure and levels of admixture for the populations. This assumption has been supported by farmers’ assumption as well about clusters on the field. The clusters shown in Figure 2 have been confirmed using BAPS program.
The genetic diversity of Burkina Faso cattle populations sampled from different regions across the country was assessed. The mean number of observed alleles was almost similar to the 11.4 alleles per locus reported by Loftus et al. (1999) but considerably higher than the 8.4 reported by MacHugh et al., (1997), 9.7 reported by Thévenon et al. (2007) in the Southern-West of Burkina Faso, 4.59 and 4.37 reported in Pakistan breeds by Rehman and Khan (2009), 7.11, 7.41 and 6.74 reported in Arabic Zebu, Bororo Zebu and Kuri cattle, respectively (Grema et al., 2017). This difference may reflect an unexpected bias in the selection of the loci but also the absence of selection pressure in cattle in Burkina Faso. In such a situation, direct comparisons may not be possible because of the markers sets and techniques used. Compared to Thévenon et al. (2007), the results were almost similar when considered only the mean number of alleles per locus from tsetse challenged area from where samples were taken as well.
In Burkina Faso, Zebu population is known to consist of Fulani Zebu, M’Bororo Zebu, Azawak Zebu originated from Niger, and a few years ago Gudali Zebu which was formally from Nigeria. Individuals from these Zebu types are thought to be included. Also, the analyses showed that a certain number of animal migrated per generation in the present populations. That is very common in diversity studies and it may be due to cattle movement along with human. In Burkina Faso, it may be due to the transhumance. The deviation in Baoulé population may
result from misclassifying N’Dama type or their cross bred in Baoulé type. N’Dama cattle were present in the South-West in the International Centre for Research and Development in Animal Husbandry in Subhumid Zones (CIRDES) research farm for scientific experiences and some individuals have been introduced in farmers’ herds. One more reason of departure from HWE could be the admixture linkage disequilibrium, the correlations that arise between linked markers in admixed populations, as described by Falush et al. (2003).
Observed and expected heterozygosity across populations were similar or comparable to those reported by Moazami-Goudarzi et al., (1997), Ibeagha-Awemu et al. (2004) in West and Central African cattle populations, Sodhi et al. (2005) in Indian cattle populations, Zerabruk et al., (2007) and Dadi et al. (2008) in Ethiopian indigenous cattle populations. But lightly different from Martin-Burriel et al. (2007) may be because the populations in the study were endangered. Average heterozygosity was within the range of 0.3 to 0.8 as suggested by Takezaki and Nei (1996) to be useful for measuring genetic variation. The overall FST revealed a moderate level of genetic differentiation among the populations in the study. The overall value of FST observed is similar to that observed in Ankole cattle in Uganda (Kugonza et al., 2010), lower than that reported in 2 Indian cattle populations (Sodhi et al., 2005) greater than that observed in Ethiopian populations (Dadi et al. 2008), in Ankole cattle in the African Great lakes region (Ndumu et al., 2008). The moderate genetic differentiation could be a result of gene flow from other populations. In the Northern part the animals are reared without any trypanosomosis pressure therefore there is less or no crossing with the taurine breed. But in the tsetse challenged regions where trypanosomosis is the most important disease in cattle (Soudre et al., 2009) crossbreeding is frequent. In addition, the pastoral production systems, long distance migrations within and across countries, utilization of communal pastures, exchange of breeding animals, uncontrolled mating facilitate constant gene flow.
Moderate genetic differentiation among indigenous cattle populations in Burkina Faso across the loci makes possible to use these breeds to improve the genetic for production and conservation and diversity in general. Regarding trypanosomosis, it may help to improve the tolerance of Zebu breed to trypanosomosis in the tsetse infested regions. Added to these advantages, little is known about the genetic diversity, structure and degree of admixture among Burkina Faso cattle populations. This supports the statement of Hanotte and Jianli (2005), knowledge of both the global diversity of the breeds and admixture events will be needed in order to be able to make sound priority decisions. Actions should be drawn to conserve the Baoulé breed which is threatened by the introgression of Zebu breed to its biotope. The Baoulé West as reported by the study cannot be differentiated from the crosses. The introgression of Zebu in the Southern areas of Burkina Faso will be perhaps more important with the climate change.
The authors have not declared any conflict of interests.
The authors are grateful to the Austrian Exchange Service (OEAD) for the grant and for funding the field work; the International Livestock Research Institute (ILRI, Kenya), the University of Natural Resources and Livestock Sciences (Boku, Austria), and the Poytechnique University of Bobo-Dioulasso (UPB, Burkina Faso) for funding the field work as well and the Veterinary Medicine University of Vienna (Austria) for the lab facilities. They also thank the farmers who allowed them to take blood samples from their animals and Dr. Abdoul Karim Ouédraogo who edited the map.
REFERENCES
|
Bradley DG, MacHugh DE, Cunningham P, Loftus RT (1996). Mitochondrial diversity and the origin of African and European cattle. Proceedings of the National Academy of Sciences of the USA 93:5131-5135.
Crossref
|
|
|
|
Bradley DG, MacHugh DE, Loftus RT, Sow RS, Hoste CH, Cunningham EP (1994). Zebu-Taurine variation in Y chromosomal DNA: a sensitive assay for genetic introgression in West African trypanotolerant cattle population. Animal Genetics 25:7-12.
Crossref
|
|
|
|
|
Caballero A, Tor MA (2002). Analysis of genetic diversity for the management of conserved subdivided populations. Conservation Genetics 3:289-299.
Crossref
|
|
|
|
|
Clutton-Brock J (1989). Cattle in Ancient North Africa. In: The Walking Larder Patterns of Domestication, Pastoralism, and Predation (ed. Clutton-Brock J), pp 200-214. Unwin-Hyman Ltd, London.
|
|
|
|
|
Corander J, Marttinen P, Sirén J, Tang J (2008). Enhanced Bayesian in BAPS software for learning genetic structure of populations. BMC Bioinformatics 9:539.
View
Crossref
|
|
|
|
|
Courtin F, Rayaissé JB, Tamboura I, Serdébéogo O, Koudougou Z, Solano P, Sidibé I (2010). Updating the Northern Tsetse Limit in Burkina Faso (1949–2009): Impact of Global Change. International Journal of Environmental Research and Public Health 7:1708-1719.
Crossref
|
|
|
|
|
Dadi H, Tibbo M, Takahashi Y, Nomura K, Hanada H, Amano T (2008). Microsatellite analysis reveals high genetic diversity but low genetic structure in Ethiopian indigenous cattle populations. Animal Genetics 39:425-431.
Crossref
|
|
|
|
|
Falush D, Stephens M, Pritchard JK (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567-1587.
|
|
|
|
|
Falush D, Stephens M, Pritchard JK (2007). Inference of population structure using multilocus genotype data: dominant markers and null alleles.
Crossref
|
|
|
|
|
Gautier M, Flori L, Riebler A, Jaffrézici F, Laloé D, Gut I, Moazami-Goudarzi K, Foulley JL (2009). A whole genome Bayesian scan for adaptative genetic divergence in West African cattle. BMC Genomics 10:550.
Crossref
|
|
|
|
|
Glaubitz JC (2004). Convert: a user friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Molecular 4:309-310.
Crossref
|
|
|
|
|
Goudet J (2002). FSTAT version 2.9.3.2: A computer program to calculate Fstatistics.
View
|
|
|
|
|
Grace D (2005). Epidemiology and control of cattle trypanosomosis in villages under risk of trypanocide resistance in West Africa. Dissertation, Freien Universität Berlin.
View
|
|
|
|
|
Grace D, Himstedt H, Sidibe I, Randolph T, Clausen PH (2007). Comparing FAMACHA eye color chart and Hemoglobin Color Scale tests for detecting anemia and improving treatment of bovine trypanosomosis in West Africa. Veterinary Parasitology 14:26-39.
Crossref
|
|
|
|
|
Grema M, Traoré A, Issa M, Hamani M, Abdou M, Soudré A, Sanou M, Pichler R, Tamboura HH, Alhassane Y, Periasamy K (2017). Short tandem repeat (STR) based genetic diversity and relationship of indigenous Niger cattle. Archives Animal Breeding 60:399-408.
Crossref
|
|
|
|
|
Guo SW, Thompson EA (1992). Performing the exact test of Hardy – Weinberg proportions for multiple alleles. Biometrics 48:361-472.
Crossref
|
|
|
|
|
Hanotte O, Jianli H (2005). Genetic characterization of livestock populations and its use in conservation decision-making. In the role of Biotechnology, Villa Gualino, Turin, held Italy, 5-7 March, 2005.
View
|
|
|
|
|
Hanotte O, Bradley DG, Ochieng JW, Verjee Y, Hill EW, Rege JEO (2002). African pastoralism: Genetic imprints of origins and migrations. Sciences 296:336-339.
Crossref
|
|
|
|
|
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009). Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources 9:1322-1332.
Crossref
|
|
|
|
|
Ibeagha-Awemu EM, Jan OC, Weimann C, Erhardt G (2004). Genetic diversity, introgression and relationship among West/Central African cattle breeds. Genetics Selection Evolution 36:673-690.
Crossref
|
|
|
|
|
Kugonza DR, Jianlin H, Nabasirye M, Mpairwe D, Kiwuwa GH, Okeyo AM, Hanotte O (2010). Genetic diversity and differentiation of Ankole cattle population in Uganda inferred from microsatellite data. Livestock science, in press.
Crossref
|
|
|
|
|
Langella O (1999). Population 1.2.30: Population genetic software (individuals or populations distances, phylogenetic trees).
View
|
|
|
|
|
Levene H (1949). On a matching problem in Genetics. Annals of Mathematical Statistics 20:91- 94.
Crossref
|
|
|
|
|
Loftus RT, Ertugrul O, Harba AH, El-Barodys MAA, MacHugh DE, Park SDE, Bradley DG (1999). A microsatellite survey of cattle from a centre of origin: the Near East. Molecular Ecology 8:2015-2022.
Crossref
|
|
|
|
|
MacHugh DE, Shriver MD, Loftus RT, Cunningham P, Bradley DG (1997). Microsatellite DNA variation and the evolution, domestication and phylogeography of taurine and zebu cattle (Bos taurus and Bos indicus). Genetics 146:1071-1086.
|
|
|
|
|
Moazami-Goudarzi K, Laloë D, Furet JP, Crosclaude F (1997). Analysis of genetic relationships between 10 cattle breeds with 17 microsatellites. Animal Genetics 28:338-345.
Crossref
|
|
|
|
|
Martin-Burriel I, Rodellar C, Lenstra JA, Sanz A, Cons C, Osta R, Reta M, De Argüello S, Sanz A, Zaragoza P (2007). Genetic diversity and relationship of endangered Spanish cattle breeds. Journal of Heredity 98(7):687-691.
Crossref
|
|
|
|
|
Ndumu DB, Baumung R, Hanotte O, Wurzinger M, Okeyo MA, Jianli H, Kibogo H, Soelkner J (2008). Genetic and morphological characterization of the Ankole longhorn cattle in the African great lakes region. Genetics Selection Evolution 40:467-490.
|
|
|
|
|
Nei M (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583-590.
|
|
|
|
|
Nei M (1987). Molecular evolutionary genetics. Columbia University Press, New York.
|
|
|
|
|
Page RDM (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12: 357-358.
View
|
|
|
|
|
Paturel JE, Servat E, Delattre MO (1998). Analyse de séries pluviométriques de longue durée en Afrique de l'Ouest et Centrale non sahélienne dans un contexte de variabilité climatique. Hydrological Sciences Journal 43:937–946.
Crossref
|
|
|
|
|
Petit RJ, El Mousadik A, Pons O (1998). Identifying populations for conservation on the basis of genetic markers. Conservation Biology 12:844-855.
Crossref
|
|
|
|
|
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics 155:945-959.
|
|
|
|
|
Rehman MS, Khan MS (2009). Genetic diversity of Hariana and Hissar cattle breed from Pakistan using microsatellite analysis. Pakistan Veterinary Journal 29(2):67-71.
|
|
|
|
|
Reynolds J, Weir BS,Cockerham CC (1983). Estimation of the coancestry coefficient: Basis for a short-term genetic distance. Genetics 105:767-769.
|
|
|
|
|
Rousset F (2008). Genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Molecular Ecology Resources 8:103-106.
Crossref
|
|
|
|
|
Sodhi M, Mukesh M, Mishra BP, Prakash B, Ahlawat SPS,Mitkari KR (2005). Evaluation of genetic differentiation in Bos indicus cattle breeds from Marathwada region of India using microsatellite polymorphism. Animal Biotechnology 16:127-137.
Crossref
|
|
|
|
|
Soudre A, Ouedraogo AG, Hanotte O, Wurzinger M, Sölkner J (2009). Trypanosomosis and cattle health management in three regions of Burkina Faso. In Tielkes, E., (ed), Proceedings of the 5th Tropentag, Hamburg 2009, (Biophysical and Socio-economic frame conditions for the sustainable management of natural resources: International research on food security, natural resource management and rural development), 244.
View
|
|
|
|
|
Takezaki PJ, Nei M (1996). Genetic distances and construction of phylogenetic tree from microsatellites DNA. Genetics 144:389-399.
|
|
|
|
|
Thévenon S, Dayo GK, Sylla S, Sidibe I, Berthier D, Legros H, Boichard D, Eggen A and Gautier M: (2007). The extent of linkage disequilibrium in a large cattle population of western Africa and its consequences for association studies. Animal Genetics 38:277-286.
Crossref
|
|
|
|
|
Whatman protocol BD01.
|
|
|
|
|
Weir B, Cockerham CC (1984). Estimation of F-statistics for the analysis of population structure. Evolutionary 38:1358-1370.
|
|
|
|
|
Yeh FC, Yang RC, Boyle T (1999). POPGENE 1.3.1: Quick user guide. University of Alberta and Center for International Forestry Research.
View
|
|
|
|
|
Zerabruk M, Benewitz J, Kantenen J, Olsaker I, Vangen O (2007). Analysis of genetic diversity and conservation priorities for six north Ethiopian cattle breeds. Journal of Animal Breeding and Genetics 124:236-241.
Crossref
|
|