ABSTRACT
The genetic diversity and relationships of 70 accessions of yam belonging to Dioscorea cayenensis/Dioscorea rotundata complex (55), Dioscorea bulbifera (13) and Dioscorea alata (2, as a reference) were assessed using six inter simple sequence repeat (ISSR) primers. DNA was extracted from a bulk of two plants per accession using a modified cetyl trimethyl ammonium bromide (CTAB) method. Six ISSR primers amplified 77 fragments with 75 (97.40%) polymorphism at genus level. The genetic diversity, estimated by gene diversity and Shannon’s index were 0.36 and 0.53, respectively, revealing a high level of genetic variation at genus level. At species level, 75 bands were amplified for D. cayenensis/D. rotundata complex, out of which 71 were polymorphic accounting for 92.2% polymorphism. Gene diversity and Shannon's index for D. cayenensis/D. rotundata complex were 0.33 and 0.49, respectively. In the case of D. bulbifera, a total of 64 bands were scored, out of which 55 were found to be polymorphic which resulted in 71.4% polymorphism. Gene diversity and Shannon's index for this species were 0.24 and 0.47, respectively. Genetic diversity analysis of D. cayenensis/D. rotundata complex accessions showed that Gedeo was the most diverse among populations and South among groups. Analysis of molecular variance (AMOVA) indicated the presence of higher proportion of variation within species (63.9%) than among species (36.1%). AMOVA for D. cayenensis/D. rotundata complex also showed higher within population variation (53.6) than among populations (46.4). In addition, cluster analysis for relationship between D. cayenensis/D. rotundata complex accessions showed grouping of some of the accessions according to their population but it failed to produce clear species boundary between D. cayenensis/D. rotundata complex. The results suggest that there is a high level of genetic diversity in Ethiopia yams to be exploited for future improvement (breeding) of the crop.
Key words: Dioscorea bulbifera, Dioscorea cayenensis/Dioscorea rotundata complex, genetic diversity, yam.
Yams are one of tuber crops which belong to the genus Dioscorea in the family Dioscoreaceae. The family is believed to be among the earliest angiosperms and probably originated from Southeast Asia (Coursey, 1967; Wilkin, 1998). Yams (Dioscorea L. spp.) are the fourth ranked and most important tuber crops in economic terms next to potatoes (Solanum tuberosum L.), cassava (Manihot esculenta Crantz) and sweet potatoes (Ipomoea batatas (L.) Poir.) (Mignouna et al., 2005). Yams (Dioscoreaspp.) are one of the most important native tuber crops of Ethiopia (EIB, 2009) and the country ranks 10th in the world in terms of their production (FAOSTAT, 2013). The cultivated species of yam, Dioscorea bulbifera, Dioscorea abyssinica and Dioscorea schimperiana are native to Ethiopia (Westphal, 1975).
The advent of the polymerase chain reaction (PCR) was a breakthrough for molecular marker techniques and made possible many fingerprinting methods. Among all markers, RAPD, inter simple sequence repeat (ISSR), amplified fragment length polymorphism (AFLP) and most recently genotyping by sequencing (GBS) markers are the most widely applied probably because they do not require the knowledge of genome sequences and the protocols used are relatively simple, rapid and cost effective (Srivastava et al., 2004; Vijayanet al., 2005; Elshireet al., 2011).
Different types of DNA molecular assays have been applied in yam in different countries (Arnau et al., 2017; Zhou et al., 2008; Nascimento et al., 2013; Muluneh et al, 2007; Wendawek et al., 2013a, b; Atnafua, 2014). However, only few studies: AFLP (Muluneh et al., 2007; Wendawek et al., 2013b) and SSR (Atnafua, 2014) have been conducted on yams of Ethiopia using molecular markers. Few studies were also conducted based on morphological and agronomic traits (Muluneh et al., 2008; Tewodros 2013). Previous researchers on yams of Ethiopia suggested the need for further studies with inclusion of areas which were not included in their studies. For instance, accessions from Ilu Ababora were not included in any of the previous studies. Moreover, D. bulbifera in Southwestern Ethiopia were not studied using any molecular markers. Besides, there is no previous report regarding the use of ISSR markers for assessment of genetic diversity of yams (Dioscorea spp.) from Ethiopia. Hence, the present study aimed at assessing the level and pattern of inter- and intra-species genetic diversity and relationships among yams (Dioscorea spp.) accessions of Ethiopia using ISSR markers. This will be of great importance to supplement actions to be taken towards improvement and conservation of the crop.
Collection of plant material
A total of 70 accessions (55 D. cayenensis/D. rotundata complex; 13 D. bulbifera; 2 D. alata) belonging to 7 populations (3 from Oromia and 4 from Southern Nations, Nationalities and Peoples Regional State, SNNPRS) were obtained from Ethiopian Institute of Biodiversity (EIB). For DNA extraction, five fresh and young leaf samples were randomly selected from each accession and silica gel dried in a zip lock bag.
DNA extraction, primer screening and PCR optimization
The ISSR marker assay was conducted at Plant Genetics Research Laboratory of the Department of Microbial Cellular and Molecular Biology, Addis Ababa University, Addis Ababa, Ethiopia. Genomic DNA was extracted following the CTAB protocol (Borsch et al., 2003). A total of 15 ISSR primers were screened for their ability to generate clear, reproducible, polymorphic and high resolution bands, in four (two from D. cayenensis/D. rotundata complex; two from D. bulbifera) randomly selected accessions. Out of 15 candidate ISSR primers, six good ones with clear, reproducible and polymorphic bands (ISSR-811, ISSR-818, ISSR-844, ISSR- 848, ISSR-873 and ISSR-880) were selected and used for analysis of genetic diversity of Dioscorea spp. (Table 1). Various combinations (at different concentrations) of PCR components were tested to find out optimum concentrations of the PCR reaction components and the one with clear band was used.
PCR amplification and agarose gel electrophoresis
PCR amplification was carried out in a 25 µl reaction mixture containing 16.7 µl sterile deionized H2O, 1 µl dNTPs (25mM each), 2.5 µl Taqbuffer (10X reaction buffer S), 2 µl MgCl2 (25mM), 0.4 µl primer (20 pmol/μl), 0.4 µl Taqpolymerase (5 unit/µl) and 2 µl diluted template DNA. Polymerase chain reactions were conducted in Biometra T3 Thermocycler withthe followingamplification program: a preheating and initial denaturation at 94°C, for 4 min, then 15 s for 40 cyclesat 94°C, 1 min primer annealing at primer annealing temperature (varies based on primers used), primer extension at 72°C for 1.30 min and final extension at 72°C for 7 min. The PCR products (10 μL for each sample) together with 1 μL 6X loading dye were loaded on 1.67% agarose gel with 100 bp ladder as a size standard (on the first lane of the gel) and subjected to electrophoresis in BIORAD min-sub®(from Biometra® cell GT at 80V standard power pack P25) for about 2 h and visualized under UV light using gel documentation system (Biosens SC750), stained by immersing in 1% ethidium bromide solution (50 µl ethidium bromide diluted with 450ml H2O) and destained with distilled H2O. The image of both stained and destained was photographed with BiosensSC750.
Statistical analysis
Amplified products were scored as present (1) or absent (0) to form a binary matrix. For analysis, scored data were subjected to different software like POPGENE (Yeh et al., 1999) to calculate genetic diversity as: number of polymorphic loci (NPL), percent polymorphic loci (PPL), Nei's gene diversity (H) and Shannon's information index (I). Areliquin (Excoffier et al., 2006) was used to compute AMOVA, while NTSYS- pc (Rohlf, 2000) and Free Tree (Pavlicek et al., 1999) were used to generate UPGMA and NJ tree, respectively. PAST & STATISTICA (Hammer et al., 2001; Statistica Soft, Inc.2001) were also used to generate two dimensional (2D) and three dimensional (3D) plots.
Banding patterns of the ISSR primers
Six ISSR primers namely 811, 818, 844, 848, 873 and 880, were selected based on the presence of well deï¬ned, informative and good resolution bands. A total of 77 bands with a size ranging from 200 to 3000 bp and an average of 13 bands per primer were obtained with six ISSR primers on 70 accession of Dioscorea spp. The highest number of scorable bands (17) was generated by penta-nucleotide ISSR-primer 880, whereas di-nucleotide ISSR-primers 844 and 811 generated the least number of scorable bands (11). The remaining ISSR-primers 818, 848 and 873 generated 13, 13 and 12 bands, respectively. Out of the six ISSR primers used, gel electrophoresis pattern obtained using primer ISSR-848 is illustrated in Figure 1.
For D. cayenensis/D. rotundata complex, a total of 75 bands were generated with number of bands produced by each primer ranging from 10 to 16, with the average bands per primer being 12. The highest number of bands was again amplified by primer 880, while primer 844 amplified the lowest number of bands (Table 2).
In D. bulbifera accessions, a total of 64 bands with 7 to 13 bands for each primer and with an average of 11 bands per primer resulted from the six ISSR primers used. Primer 880 and 818 produced the highest number of bands, while the lowest number of bands was amplified by primer 811 (Table 2).
Species specific ISSR bands in D. cayenensis/D. rotundata complex and D. bulbifera
Out of the six ISSR primers used, five ISSR primers showed species specific bands (Table 2). A total of 13 bands specific to D. cayenensis / D. rotundatacomplex were generated. Primers 811 and 880 showed the highest number of specific bands to this species (four bands). Only two bands specific to D. bulbifera were amplified by six primers. Two bands specific to D. bulbifera were generated by primers 844 and 880. ISSR primer 818 generated no specific bands for both species whereas primers 811, 848 and 873 generated specific bands for only D. cayenensis/D. rotundatacomplex (Table 2).
Application of ISSR markers in Dioscorea species genetic diversity assessment
Six ISSR primers amplified 77 fragments with 75 (97.40%) polymorphism at genus level. The genetic diversity, estimated by Gene diversity and Shannon’s index were 0.36 and 0.53, respectively, revealing a high level of genetic variation at genus level. At species level, 75 bands were amplified for D. cayenensis/D. rotundata complex, out of which 71 were polymorphic accounting for 92.2% polymorphism. Gene diversity and Shannon's index for D. cayenensis/D. rotundata complex were 0.33 and 0.49, respectively. Comparable results have been reported by Wendawek et al. (2013b) who used AFLP fingerprinting to evaluate and characterize 43 individuals belonging to different populations of wild and cultivated guinea yam (D. cayenensis/D. rotundata complex) using three-primer combination and detected 78% polymorphism.
Bressan et al. (2014) also evaluated 21 local varieties of D. cayenensis and two D. rotundata accessions using 7 isozyme loci and 24 morphological markers, and reported the existence of high genetic variability with 100% polymorphism using isozyme marker. Dansi et al. (2000) and Mignougna et al. (2002) also studied genetic diversity of D. cayenensis/D. rotundata complex using isozyme markers in 7 and 6 isozyme systems, respectively, and reported the existence of high diversity (polymorphism in all analyzed isozyme systems) which is in agreement with the present study.
In the case of D. bulbifera, a total of 64 bands were scored, out of which 55 were polymorphic which resulted in 71.4% polymorphism. Gene diversity and Shannon's index for this species were 0.24 and 0.47, respectively. Despite its smaller sample size (13), D. bulbifera accessions still showed high genetic diversity but lower than D. cayenensis/D. rotundata complex. This shows that small populations or individuals are not always associated with a lack or low level of genetic variation (Yingjuan and Ting, 2009). Likewise, Tewodros (2013) studied the level of genetic diversity within D. bulbifera accessions collected from South and Southwestern Ethiopia based on key agronomic traits and reported the existence of high diversity in the region. Silva et al. (2016) also evaluated genetic diversity among 42 D. bulbifera accessions from Brazil using microsatellite markers and found high genetic diversity.
Both D. cayenensis/D. rotundata complex and D. bulbifera showed high genetic diversity, 92.2 and 71.4% at the species level, respectively. Shannon's information index of both species (0.49 for D. cayenensis/D. rotundata complex; 0.47 for D. bulbifera) was also higher than the average values for widespread species (0.202) as suggested by Hamrick and Godt (1989).
Among the two species, D. cayenensis/D.rotundatacomplex was more diverse, this might be due to its larger sample size, being a combination of various populations and for representing two species complex. Genetic diversity analysis of D. cayenensis/D. rotundata complex populations showed that Gedeo was the most diverse, while East Wellega was the least diverse. Gedeo areas are known for their traditional agro-forestry system in which they grow a variety of crop plants including tuber crops (Wubalem, 2014), which might be the reason for the highest diversity of yams in the area. In the case of East Wellega, it is geographically a bit isolated with no/little incoming genes or tubers as a seed. Therefore, it is more likely that the genetic diversity in this area might be lower due to presence of lower or no tuber exchange with other populations.
Partitioning of genetic diversity in yams (Dioscorea spp.)
AMOVA revealed the presence of higher proportion of variation within species (63.9%) than among species (36.1%) (Table 3).This might be due to the presence of several shared bands between these species, which indicates that they might have close evolutionary relationship and/or admixture on farmer’s field might have facilitated gene flow (pollen flow). It has been reported that D. cayenensis/D. rotundata complex and D. bulbifera are most likely cross compatible due to their similar ploidy level (2n= 40, 60, 80) (Coursey, 1967; Asiedu, 1997). The potential for gene exchange has long been recognized even between taxa with large differences in chromosome numbers (Stebbins, 1971).
AMOVA for D. cayenensis/D. rotundata complex also showed moderately higher within population variation (53.6) than among populations (46.4) (Table 4). Similarly, Similarly, Loko et al. (2016) used microsatellite marker to study genetic diversity and relationship of guinea yam germplasm of Benin and found 96% of variation within population and 4% among population. Muluneh et al. (2007) also assessed genetic diversity of yam (Dioscorea spp.) germplasms from Ethiopia and their relatedness to the main Dioscorea spp. by AFLP markers and found 81% of the total genetic variation being attributed to within populations and only 19% to among populations. Wendawek et al. (2013a) also evaluated genetic diversity and population structure of guinea yams and their wild relatives in South and South West Ethiopia using microsatellite marker, reporting the same pattern. This could be due to gene flow through seed materials exchange among local farmers.
Genetic relationships within and among species
Cluster analysis showed grouping of most of the accessions according to their species. Mignouna et al. (2005) also used RAPD and double stringency PCR (DS-PCR) and reported similar result. In addition, cluster analysis for relationship between D. cayenensis/D. rotundata complex accessions showed grouping of some of the accessions according to their population. Accessions from East Wellega clearly formed their own cluster, accessions from Semen Omo were grouped together with those of Hadiya-Kembata, while accessions from Gedeo were clustered together with those of Jimma (Figure 2). Both of the two (Figure 3a) and three dimensional PCO plots (Figure 3b) showed the same pattern. Similarity between Semen Omo and Hadiya-Kembata population is expected due to geographical proximity of those areas. However, genetic similarity was also present between Gedeo and Jimma accessions in spite of their geographical distance. Hence, this study showed that there is no strong correlation between geographic distance and genetic diversity. This could be explained in terms of movement of the people carrying tubers and distribution of cultivars over great distance as clones in the course of human movement.
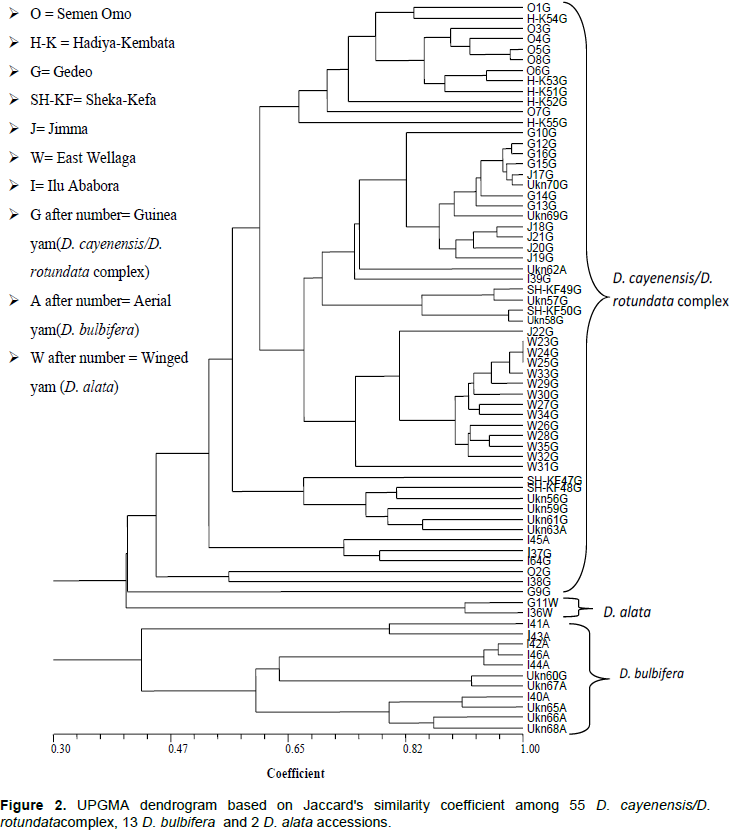
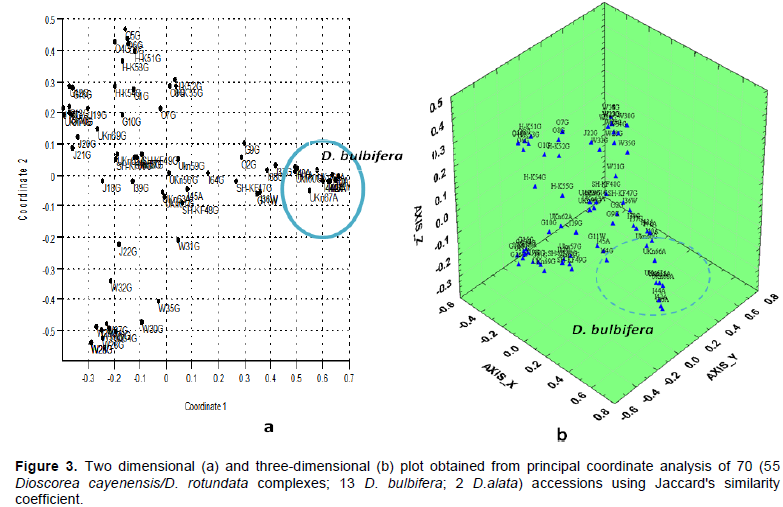
ISSR data failed to produce any clear boundary between different types of Guinea yams that showed domestication characteristics of different species (wild, cultivated and intermediate) based on their tuber flesh colour. Similarly, Wendawek et al. (2013b) used AFLP genetic finger printing to evaluate and characterize 43 individual plants belonging to different populations of wild and cultivated guinea yams and reported that ordination and cluster analysis did not produce any clear boundary between either the guinea yam accessions or between them and their wild relatives. The finding of the present ISSR data failed to produce any clear boundary between different types of Guinea yams that showed domestication characteristics of different species (wild, study supports the reports of Miege and Sebsebe (1997), which indicates that they are species complex with many intermediates.
Both D. cayenensis/D. rotundata complex and D. bulbifera showed high genetic variation. In D. cayenensis/D. rotundata complex, the highest genetic diversity was found within Gedeo population, which indicates that this population can be considered as a source of diverse individuals in future improvement of the crop. On the contrary, East Wellega population, which showed the least diversity, needs special attention for conservation. Variation within species seemed to be greater than that of among species. Similarly, AMOVA analysis of D. cayenensis/D. rotundata complex populations showed higher within population variation than among population variation which indicates existence of high level of gene flow. Cluster and PCO analyses showed clustering of most of the accessions to their respective species and in some cases, to their geographic origin. However, they failed to differentiate between different guinea yam (D. cayenensis/D. rotundata complex) types, which support the idea that they are species complex.
The authors have not declared any conflict of interests.
REFERENCES
|
Arnau G, Bhattacharjee R, MN S, Chair H, Malapa R, Lebot VKA, Perrier X, Petro D, Penet L, Pavis C (2017). Understanding the genetic diversity and population structure of yam (Dioscorea alata L.) using microsatellite markers. PLoS one 12(3):e0174150.
Crossref
|
|
|
|
Asiedu R, Wanyera NM, Ng SY, Ng NQ (1997).Yams. In: Fuccillo D, Sears L. and Stapelton P(eds.) Biodiversity in trust: conservation and use of plant genetic resources in CGIAR centers. Cambridge University Press, Cambridge, UK, pp. 57-66.
Crossref
|
|
|
|
|
Atnafua B (2014). Studies on Molecular Genetic Diversity and Useful Genomic Traits of Yam (Dioscorea Spp.) Germplasm Collections from Ethiopia.PhD Thesis, Addis Ababa University, Addis Ababa.
|
|
|
|
|
Borsch T, Hilu KW, Quandt D, Wilde V, Neinhuis C,Barthlott W (2003). Noncoding plastid trnT-trnFsequences reveal a well resolved phylogeny of basal angiosperms. Journal of Evolutionary Biology 16:558-576.
Crossref
|
|
|
|
|
Bressan EA,Neto TB, Zucchi MI, Rabello RJ,Vease EA (2014).Genetic structure and diversity in the D. cayenensis/D. rotundata complex revealed by morphological and isozyme markers. Genetics and Molecular Research 13(1):425-437.
Crossref
|
|
|
|
|
Coursey DG (1967). Yams: An Account of the Nature, Origins, Cultivation and Utilisation of the Useful Members of the Dioscoreaceae. London.
View
|
|
|
|
|
Dansi A, Mignouna HD, Zoundjihekpon J, Sangare A, Asiedu R, Ahoussou N (2000).Using isozyme polymorphism to assess genetic variation within cultivated yams (Dioscoreacayenensis/Dioscorearotundata complex) of the Republic of Benin. Genetic Resources and Crop Evolution 47:371-383.
Crossref
|
|
|
|
|
Ethiopian Institute of Biodiversity (EIB) (2009). Ethiopia's 4th Country Report. Convention on Biological Diversity, Addis Ababa, Ethiopia.
View
|
|
|
|
|
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379.
Crossref
|
|
|
|
|
Excoffier L, Laval G. and Schneider S (2006)An Integrated Software Package for Population Genetics Data Analysis. Computational and Molecular Population Genetics Lab (CMPG), Institute of Zoology, University of Berne, Switzerland.
View
Crossref
|
|
|
|
|
FAOSTAT (2013). 2010-2013 Factfish, Germany.
View.
|
|
|
|
|
Hammer O, Harper DA and Ryan PD (2001). PAST: Paleontological statistics software package for education and data analysis. Palaeo electronic4:9.
View.
|
|
|
|
|
Hamrick J and Godt M (1989) Allozyme diversity in plants In: Brown AH, Clegg MT, Kahler AL and Weir BS (eds.) Plant Population Genetics, Breeding and Genetic Resources, Sinauer, Sunderland P 123.
|
|
|
|
|
Loko YL, Bhattacharjee R, Agre AP, Dossou-Aminon I, Orobiyi A, Djedatin GL, Dansi A (2016). Genetic diversity and relationship of Guinea yam (Dioscorea cayenensis Lam.-D. rotundata Poir. complex) germplasm in Benin (West Africa) using microsatellite markers. Genet. Genetic Resources and Crop Evolution pp. 1-15.
Crossref
|
|
|
|
|
Mignouna HD, Abang MM, Wanyera MW, Chikaleke VA, Asiedu R,Thottappilly G (2005). PCR marker-based analysis of wild and cultivated yams (Dioscorea spp.) from Nigeria: genetic relationships and implications for ex situ conservation. Genetic Resources and Crop Evolution 52:755-763.
Crossref
|
|
|
|
|
Muluneh T, Heiko CB and Brigitte LM (2008). Diversity, distribution and management of yam landraces (Dioscorea spp.) in Southern Ethiopia. Genetic Resources and Crop Evolution 55:115-131.
Crossref
|
|
|
|
|
Muluneh T, Heiko CB, Brigitte LM (2007). Genetic diversity in yam germplasm (Dioscorea spp.) from Ethiopia and their relatedness to the main cultivated Dioscorea species assessed by AFLP markers. Crop Science 47:1744-1753.
Crossref
|
|
|
|
|
Nascimento WF, Rodrigues JF, Kochier S, Gepts P, Veasey EA (2013). Spatially structured genetic diversity of the Amerindean yam (Dioscorea trifida L.) assessed by SSR and ISSR markers in Southern Brazil. Genetic Resources and Crop Evolution 60:2405-2420.
Crossref
|
|
|
|
|
Pavlicek A, Hrda S, Flegr J (1999). Free-tree-software program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness.Application in the RAPD analysis of genus Frenkelia. Folia Biologica (Praha). 45:97-99.
|
|
|
|
|
Rohlf FJ (2000). NTSYS-pc ver 2.11T.Exter Software, Setauket, New York.
|
|
|
|
|
Silva DM, Siqueira MV, Carrasco NF, Mantello CC, Nascimento WF, Veasey EA (2016). Genetic diversity among air yam (Dioscorea bulbifera) varieties based on single sequence repeat markers. Genetics and Molecular Research 15(2):gmr.15027929.
Crossref
|
|
|
|
|
Srivastava PP, Vijayan K, Awasthi AK, Saratchandra B (2004). Genetic analysis of Morusalba through RAPD and ISSR markers. Indian Journal of Biotechnology 3:527-532.
|
|
|
|
|
Statistica Stat Soft, Inc.2001. STATISTICA (data analysis software system) Version 6.0.
|
|
|
|
|
Stebbins GL (1971). Chromosomal Evolution in Higher Plants. Edward Arnold LTD, London pp. 87-89.
|
|
|
|
|
Tewodros M (2013).Genetic diversity of aerial yam (Dioscoreabulbifera (L.)) accessions in Ethiopia based on agronomic traits. Agriculture Forestry and Fisherie 2(2):67-71.
Crossref
|
|
|
|
|
Vijayan K, Nair CV, Kar PK, Mohandas TK, Saratchandra B,RajeUrs S (2005).Genetic variability within and among three ecoraces of the tasar silkworm Antheraeamylitta Drury, as revealed by ISSR and RAPD markers. International Journal of Industrial Entomology 10(1):51-59.
|
|
|
|
|
Wendawek A, Sebsebe D, Fay MF, Smith RJ, Nordal I, Wilkin P (2013a). Genetic diversity and population structure of Guinea yams and their wild relatives in South and South West Ethiopia as revealed by microsatellite markers. Genetic Resources and Crop Evolution 60:529-541.
Crossref
|
|
|
|
|
Wendawek A, SebsebeD, Fay MF, Smith RJ, Nordal I, Wilkin P (2013b).Genetic diversity and species delimitation in the cultivated and wild Guinea yams (Dioscorea spp.) from Southwest Ethiopia as determined by AFLP (amplified fragment length polymorphism) markers. Genetic Resources and Crop Evolution 60:1365-1375.
Crossref
|
|
|
|
|
Westphal E (1975).Agricultural systems in Ethiopia. Center for Agriculture Publishing and Documentation, Wageningen, The Netherlands, p 278. http://edepot.wur.nl/361350
|
|
|
|
|
Wilkin P (1998).A morphometric study of Dioscoreaquartiniana. A. Rich (Dioscoreaceae). Kew Bulletin 54:1-18.
Crossref
|
|
|
|
|
Wubalem T (2014).Gedeo multistory agroforestry system and konso agroforestry and cultural landscape.GIAHS steering and scientific commitee meeting Rome, Italy.
|
|
|
|
|
Yeh F, Yang R, Boyle, T (1999).Population genetic analysis of codominant markers and qualitative traits. Belgian Journal of Botany 129:157.
|
|
|
|
|
Yingjuan S, Ting W (2009).High ISSR variation in 14 surviving individuals of Euryo Dendron excelsum (Ternstroemiaceal) endemic to china. Biochemical Genetics 47:56-65.
Crossref
|
|
|
|
|
Zhou Y, Zhou C, Yao H, Liu Y, Tu R (2008).Application of ISSR markers in detection of genetic variation among Chinese yam (Dioscorea opposite Thunb) cultivars. Life Science Journal 5(4):6-12.
|
|