ABSTRACT
The advent and application of molecular markers technology enabled dissection of genomic regions underling important agronomic traits in crop plants. Isolation of sufficient quantity and good quality DNA from a large number of plant samples is a major challenge in most genetic and genomic analyses. Wheat is the world’s third most important food security crop on which most molecular breeding and genetic engineering researches have been undertaken. So far, several wheat DNA isolation protocols and commercial kits are available. However, in some, the cost is so prohibitive for small laboratories, and hence finding cost effective alternative makes small biotechnology laboratories of developing countries beneficial to the technology. The present study was targeted to provide alternative economical, reliable and safer genomic DNA isolation method in wheat. Accordingly, genomic DNA was isolated from 27 bread wheat recombinant inbred lines (RIL) raised in greenhouse with and without liquid nitrogen. From 100 mg, 20-day old leaf samples, averagely 1890.7 ng/μl sufficiently pure genomic DNA (A260/A280 ratio of 1.8) was extracted. In the extraction process, ammonium acetate and sodium acetate solutions were also avoided. Polymerase chain reaction (PCR) analysis of the extracted DNA using two simple sequence repeats (SSR) markers produced polymorphic bands confirming the suitability of the extracted DNA for PCR. Therefore, it is possible to deduce that we have adopted a simple, cost effective, reliable and safer DNA isolation method that can help to extract good quality and sufficiently pure genomic DNA for further molecular studies.
Key words: Wheat, genomic DNA isolation, liquid nitrogen, polymerase chain reaction (PCR) analysis, simple sequence repeats (SSR) markers.
Nowadays, molecular markers are widely used to promote the speed and efficiency of crop improvement program. Molecular marker technology becomes a very useful tool toidentify and localize genes/Quantitative Trait Loci (QTLs) underling economically important agronomic traits, germplasm characterization, genetic diagnostics, characterization of transformants, study of genome organization, phylogenetic analysis, marker-assisted selection (Gupta et al., 1999). So far, several DNA marker systems have been developed and are routinely being applied in plant breeding program. Some of them are restriction fragment length polymorphsm (RFLPs), random amplified polymorphic DNAs (RAPD), amplified fragment length polymorphism (AFLP), inter-simple sequence repeat (ISSR), simple sequence repeats (SSR) or microsatellite and single nucleotides polymorphism (SNPs).Although each marker system has its own pros and cones, next generation sequence derived SNPs marker has been considered to be the most powerful, automated and abundant marker system suitable for genomic studies on a wide range at genomic scales (Rafalski, 2002; Zhu et al., 2003).
DNA extraction is the preliminary step in all DNA marker assays. Marker assisted crop improvement involving scanning of large sets might be hundreds to thousands of germplams, and isolation of sufficient quantity and high quality genomic DNA from such huge plant samples is the major bottleneck in most genetic and genomic studies (Xin and Chen, 2012). Although the advent of polymerase chain reaction (PCR) enabled to amplify minute quantity (nano-gram) of DNA, production of reproducible PCR product requires sufficiently pure DNA. Wheat is the world’s third most important food security crop (Green et al., 2012) on which most molecular breeding and genetic engineering researches have been undertaken.
So far, enormous wheat DNA isolation protocols are available including Stein et al. (2001), Dellaporta et al. (1983), Sharp et al. (1988), Murray et al. (1980), Chun et al. (1993), McCarthy et al. (2002), Clarke et al. (1989), Benito et al. (1993), etc. Including these, most reported plant DNA isolation protocols utilize liquid nitrogen for easily grinding of plant tissues without degradation of the intact DNA by the plant’s native enzymes. However, liquid nitrogen has been complained to be hazardous, difficult for handling and costly (Saini et al., 2013). In addition, although several plant genomic DNA extraction commercial kits are available (Xin and Chen, 2006); their cost is so prohibitive for small laboratories (Xin and Chen, 2012).Hence, adopting or developing cost effective DNA isolation method which can eliminate the requirement of liquid nitrogen or highly costly DNA isolation kit can be promising and economical to scientists of developing world. Therefore, the present research was initiated to develop/adopt an economical genomic DNA isolation protocol from wheat fresh leave samples without liquid nitrogen for downstream molecular studies.
The experiment was conducted at International Center for Genetic Engineering and Biotechnology (ICGEB), New Delhi, India in 2016. As part of the protocol, 100 mg of bread wheat leaves were collected from 20 days old seedlings raised under greenhouse condition. Collected leaves were rinsed with distilled water, blot dried, cut into1 to 1 – 2 cm length and placed in sterile mortar and pastel. After rapping with aluminum foil, the mortar and pestle (with the sample) were maintained in -70°C freezer for 12 h. Then, deep frozen leaves were quickly gridded using mortar and pestle and transferred into sterile 2.0ml Eppendorf tube using sterile spatula. Subsequently, 700 μl pre-warmed (65°C) 2% CTAB extraction buffer (composed of 1 M Tris-HCl, 5 M NaCl, 0.5 M EDTA, 2.0 g CTAB powder and 1.0 g PVP) with 0.2 vol% β-mercaptoethanol was added and then incubated in water bath at 65°C for 40 min. Then, 800 μl of chloroform/ iso-amayl alcohol (24:1) was added and shaken vigorously. After centrifugation at 12,000 rpm for 10 min, the supernatant (~ 600μl) was transferred into a new 1.5 ml tube. After adding 600μl cold 100% iso-propanol, it was carefully shaken by inversing the Eppendorf cap. Freezing for 2 h at -20°C followed by centrifugation at 12,000 rpm for 10 min resulted in DNA pellet. The DNA pellet was washed twice with 800 μl of cold 75% ethanol with subsequent centrifugation at 12,000 rpm for 10 min. The resulted DNA pellet was air-dried and re-suspended in 100 μl of 1× TE buffer for 1 h at room temperature. Finally, 1 μl of RNaseA (10 mg/ml) was added and incubated in water bath at 37°C for 30 min. The resulted DNA concentration and quality were determined using Nanodrop Spectrophotomer at wavelength of A260and A280 and in 1% agarose gel at 100V for 1 h. Nanodrop readings of the DNA isolated with and without liquid nitrogen were analyzed using SAS software 9.2 version (SAS Institute Inc., 2008).
Polymerase chain reaction (PCR) test
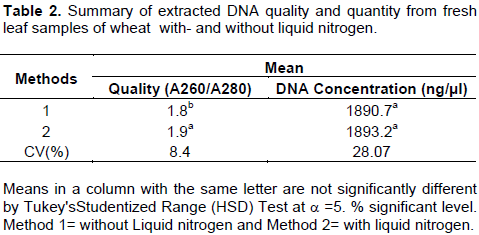
The DNA obtained the current protocol was tested using available and published two polymorphic SSR markers closely linked to Septoria tritici blotch resistance gene stb 6 (Figure 1A) and stb11 (Figure 1B) in wheat. The PCR amplification was carried out in 25 μl reaction mixture that composed of 10×PCR buffer with MgCl2,10 mM dNTPs, 0.4 μM each of forward and reverse primers, 1 μl dimethyl sulfoxide (DMSO), 1 μl Taq DNA polymerase (5 Units), 1 μl template DNA and 16.5 μl double distilled sterile H2O using a thermal cycler (Applied Biosystems Veriti 96 Well Thermal Cycler). Amplification conditions involved an initial denaturation at 94°C for 3 min followed by 45 cycles with 1 min denaturation at 94°C, 1 min annealing at 55°C (xgwm369) and 52°C (xbarc137), primer extension at 72°C for 2 min followed by final extension step of 10 min at 72°C. PCR product was fractionated in 3% agarose gel electrophoresis using 1× TAE buffer at 100V for 2 h. The gel was stained with ethidium bromide and visualized under UV light and subsequently photographed. A 50 bp ladder was used to determine the amplification size.
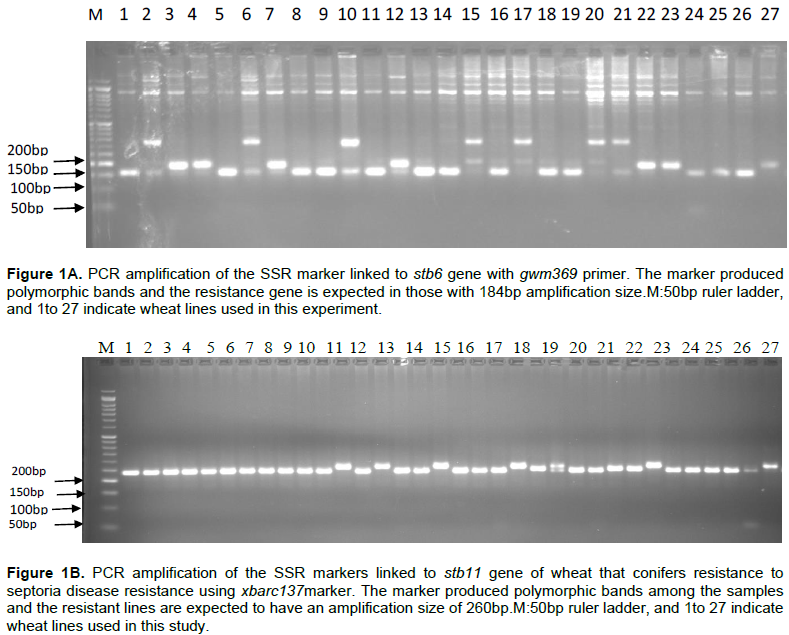
To determine the reliability and efficiency of the current extraction method, genomic DNA was isolated from 27 bread wheat RIL obtained from CIMMYT both with- and without liquid nitrogen (Table 1). As shown in Tables 1 and 2, the quality of extracted DNA estimated based on UV absorbance ratio of λ260 to λ280 is averagely 1.8458 (Table 2) which lies in the acceptance range of 1.8 to 2.0. It was confirmed that the DNA extracted in the present method without liquid nitrogen is sufficiently pure for further molecular downstream studies. Surprisingly, this quality is significantly higher than the DNA extracted using liquid nitrogen. This might be due to deep freezing of the mortar and pestle which provided longer cold environment to facilitate easy crushing of the leave samples and to inhibit DNA degradation by plant native enzymes.
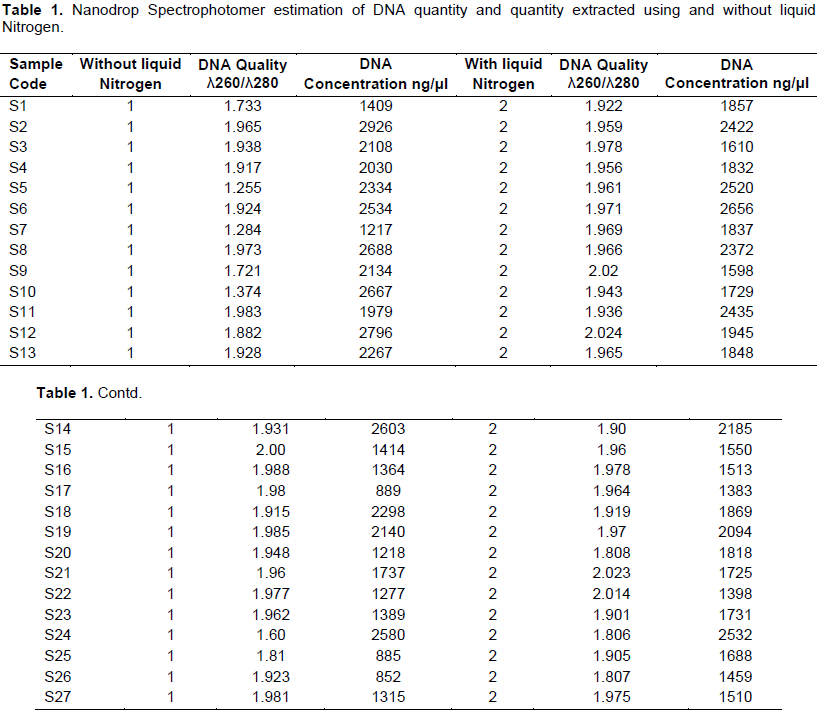
Regarding extracted DNA concentration, there is no statistically significant difference by Tukey's Studentized Range (HSD) Test at a = 5% significant level (Table 2). In PCR reaction, the amount of DNA concentration required per reaction is small (about 100ng). Thus, the DNA concentration produced by the present protocol is sufficiently enough for 1890 reactions. Hence, it is possible to deduce that the current protocol is sufficiently reliable in producing high quantity of genomic DNA that can be used for marker assisted selection (MAS), mutant analysis and other genomic studies that involve large sample studies. The very interesting finding of this research is not only avoiding the use of liquid nitrogen, it also does not involve the use of Proteinase K, ammonium acetate and sodium acetate which are commonly used by other DNA isolation protocols (Dellaporta et al., 1983; George, 2004; Dehestani and Tabar, 2007; Shahriar et
al., 2011).
The advent and application of molecular marker technology enabled plant scientists to track the transfer or presence of gene/QTLs underlining useful agronomic traits in crop plants. Most genetic and genomic studies like MAS, mutant analysis in transformation and diversity analysis involve screening of large sets of samples, and preparation of good quality and sufficient amount of DNA from such huge plant samples is the major challenge. So far reported DNA isolation protocols Such as CTAB (Saghai-Maroof et al., 1984), SDS (Sodium Dodesyl Sulfate) (Edwards et al. 1991) and other protocols (Dellaporta et al., 1983; Rogers and Benedich, 1985; Doyle and Doyle, 1990; Ziegenhagen and Scholz, 1993; Lodhi et al., 1994) used liquid nitrogen for easily grinding of plant tissues and to deactivate the plant’s native enzymes to avoid DNA degradation. However, liquid nitrogen is compiled to be hazardous, difficult to handle and costly (Saini et al., 2013). Hence, the present study was targeted at designing a simple, economical and safe genomic DNA isolation method in wheat without the use of liquid nitrogen. Accordingly, 27bread wheat RILs were used in the study. Using 100 mg leaf samples, averagely 1890.7 ng/μl good qualities (A260/A280 ratio of 1.8) genomic DNA was isolated. This interesting result was obtained not only by avoiding liquid nitrogen but also other reagents like Proteinase K (the most costly); ammonium acetate and sodium acetate solutions were not used in this improved method. The isolated DNA samples were checked with PCR using two SSR markers linked to stb genes responsible for septoria resistance. Interestingly, the two markers amplified in all the 27 samples, and produced polymorphic bands. Therefore, it is important to deduce that we have produced a simple, reliable and economical DNA isolation method that does not depend on liquid nitrogen, Proteinase K, ammonium acetate and sodium acetate which fevers small laboratories in developing countries.
The authors have not declared any conflict of interests.
REFERENCES
|
Benito C, Figueiras AM, Zaragoza C, Gallego FJ, Pena A (1993). Rapid identification of Triticeae genotypes from single seeds using the polymerase chain reaction. Plant Mol. Biol. 21:181-183.
Crossref
|
|
|
|
Chun WJ, Martin GB, Tanksley SD (1993). Pre-germination genotypic screening using PCR amplification of half-seed. Theor. Appl. Genet. 86:694-698.
|
|
|
|
|
Clarke BC, Moran LB, Appels R (1989). DNA analysis in wheat breeding. Genome 32:334-339.
Crossref
|
|
|
|
|
Dehestani A, Tabar SKK (2007). A Rapid Efficient Method for DNA Isolation from Plants with High Levels of Secondary Metabolites. Asian J. Plant Sci. 6:977-981.
Crossref
|
|
|
|
|
Dellaporta SL, Wood J, Hicks JB (1983). A plant DNA minipreparation version 2. Plant Mol. Bio. Rep. 1:19-22.
Crossref
|
|
|
|
|
Doyle JJ, Doyle JJ (1990). A rapid DNA isolation procedure for fresh plant tissue. Focus 12:13-15.
|
|
|
|
|
Edwards K, Johnstone C, Thompson C (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acid Res 19:1349.
Crossref
|
|
|
|
|
George SM (2004). A Simple Extraction Method Suitable for PCR Based Analysis of Plant, Fungal, and Bacterial DNA. Plant Mol. Biol. Rep. 22:71-81.
Crossref
|
|
|
|
|
Green AJ, Berger G, Griffey CA, Pitman R, Thomason W, Balota M, Ahmed A (2012). Genetic yield improvement in soft red winter wheat in the Eastern United States from 1919 to 2009. Crop Sci. 52:2097-2108.
Crossref
|
|
|
|
|
Gupta PK, Varshney RK, Sharma PC, Ramesh B (1999). Molecular markers and their applications in wheat breeding. Plant Breed. 118:369-390.
Crossref
|
|
|
|
|
Lodhi MA., Ye GN, Weeden NF, Reisch BI (1994). A simple and efficient method for DNA extraction from grapevine cultivars, Vitis species and Ampelopsis. Plant Mol. Biol. Rep. 12(1):6-13.
Crossref
|
|
|
|
|
McCarthy PL, Hansen JL, Zemetra RS, Berger PH (2002). Rapid identification of transformed wheat using a half-seed PCR assay prior to germination. Biotechniques 32:560-564.
|
|
|
|
|
Murray MG, Thompson WF (1980).Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res. 8(19):4321-4325.
Crossref
|
|
|
|
|
Rafalski A (2002). Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 5:94-100.
Crossref
|
|
|
|
|
Rogers SO, Benedich AJ (1985). Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 5:69-76.
Crossref
|
|
|
|
|
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984). Ribosomal DNA spacer-length polymorphism in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 81:8014-8019.
Crossref
|
|
|
|
|
Saini P, Aneja B, Yadav NR, Yadav RC (2013). Isolation of genomic DNA from leaf samples of Indian mustard without liquid nitrogen for use in molecular marker analysis. Crop Improv. 40(1):30-33.
|
|
|
|
|
SAS Institute Inc. (2008). SAS/STAT ® 9.2 User's Guide. Cary, NC: SAS Institute Inc.
|
|
|
|
|
Shahriar M , Haque Md. R, Kabir S , Dewan I, Bhuyian MA (2011). Effect of Proteinase-K on Genomic DNA Extraction from Gram-positive Strains. Stamford J. Pharm. Sci. 4(1):53-57.
Crossref
|
|
|
|
|
Sharp PJ, Kreis M, Shewry PR, Gale MD (1988). Location of β-amylase sequences in wheat and its relatives. Theor. Appl. Genet. 75:286-290.
Crossref
|
|
|
|
|
Stein N, Herren G, Keller B (2001). A new DNA extraction method for high-throughput marker analysis in a large genome species such as Triticum aestivum. Plant Breed. 120:354-356.
Crossref
|
|
|
|
|
Xin Z, Chen J (2006). DNA Sequencing II: Optimizing Preparation and Cleanup. In. Extraction of Genomic DNA from Plant Tissue. Edited by Sudbury KJ. MA: Jones and Bartlett Publishers; 2006:47-59.
|
|
|
|
|
Xin Z, Chen J (2012). A high throughput DNA extraction method with high yield and quality. Plant Methods 8:26.
Crossref
|
|
|
|
|
Zhu YL, Song QJ, Hyten DL, Van Tassell CP, Matukumalli LK, Grimm DR, Hyatt SM, Fickus EW, Young ND, Cregan PB (2003). Single-nucleotide polymorphisms in soybean. Genetics 163:1123-1134.
|
|
|
|
|
Ziegenhagen B, Scholz F (1993) A procedure for mini-preparation of genomic DNA from needles of silver fir (Abies alba Mill.). Plant Mol. Biol. Rep. 11:117-121.
Crossref
|
|