ABSTRACT
Correct identification of plants is a prerequisite to achieving desirable results in health care delivery, sustainable food production and housing, forest resources management and environmental protection. However, many of the paper-based/printable taxonomic key formats available to the taxonomist for this important responsibility are fraught with inadequacies some of which include fixed sequence of plant identification steps, non- or hardly-susceptible to computerisation, lack of provision for confirmation of suspected plant identity and indeterminable character states, and tedious construction and navigation procedures. This paper with the aim of making the practice of plant taxonomy more attractive, less laborious and dreaded, proposes two new key formats with highlights of their design/features, construction procedures and usage. These alternative key formats, with varying capacities to circumvent some of the enumerated challenges are multi-level table of identification and multi-layer circular diagnostic chart. The status of each of the proposed key formats is discussed with reference to the inadequacies observed in the dichotomous key format with which most taxonomists are familiar. Based on their structural features and functionality attributes, it is conclusive that the two alternative key formats constitute useful templates upon which reliable plant diagnostic tools can be based.
Key words: Automated plant identification, computerised key, diagnostic key, dichotomous key, multi-access key, plant identification, single-access key, taxonomic key.
The basic terminology in systematics is ‘taxon’, a formal category of living things, or a ‘taxonomic group’ recognised by having certain characteristics in common which we take as evidence of genetic relationship, and possessed some degree of objective reality (Rickett, 1958). It is a group of one or more individuals, or of lower taxa judged sufficiently similar to each other to be treated together formally as a single evolutionary or informational unit at a particular level in the taxonomic hierarchy, and sufficiently different from other groups of the same rank to be treated separately from them (Radford et al., 1974). Going by these regular definitions, each taxon (except the highest), such as a species belongs to one and only one taxon of the next higher rank, such as a genus, implying each individual belongs to exactly one species (and has one name) in any particular taxonomic treatment of its group. Taxonomists frequently present organised written descriptions of the characteristics of similar taxa such as species or genera, etc., to facilitate identification or recognition of unknown organisms. These organised descriptions are referred to as diagnostic keys, which come in different formats and styles such as in Hopkins and Stanfield (1966), Lowe and Stanfield (1974), Payne et al. (1974), Payne and Preece (1980), Jones et al. (1998) and Javatpoint (2018), each with its merits and demerits.
Outside the understanding of the terminology from the systematics (strict) point of view, and for all practical purposes, it is useful to operationally define ‘taxon’ as one or more objects recognized by sharing certain characteristics, and representing a group or category, the members of which may or may not be related taxonomically. Such objects will include tangible items as plant specimens of one or more taxonomic groups (e.g. species, sub-genus, genus, etc.), plant organs (e.g. root, flower, seed, fruit, etc.), or intangibles and other forms of categorisation such as plant diseases, colours, sounds, odours and so on (Amante and Norton, 2003). So, in the context of this study, taxon is more widely viewed beyond its conventional usage as ‘taxonomic group’ or assemblage of plants or animals that are genetically related, or so related to the best of our knowledge (Rickett, 1958) to mean a unit of classification (taxonomic or otherwise) of objects (tangible or intangible), recognised by a set of features that distinguish (or diagnose) the group from such other groups. This position equally recognises both the relationship of inclusion between levels and of complementarity within levels as aptly described by Price (1967) regarding the objects classified into groups under groups. That is, given a large group of taxa (or objects), and based on the relationships among the objects, recursive classification or compartmentalisation of the group members into taxa of lower categories/ranks is possible until the entire group is resolved into the smallest manageable clusters of taxa, or each taxon as it were.
Correct identification of plants is important in health care delivery (Upton and Romm, 2010), sustainable food production (Amante and Norton, 2003) and housing, criminal justice (Bock and Norris, 2016), forest resources management and environmental protection. Medicinal plants misidentification and misrepresentation are two known root causes of herb adulteration or substitution, which in turn, is the basic cause of serious health problems to consumers of herbal medicinal products (Panter et al., 2014), and a motivation for bad publicity and legal burdens sometimes faced by the pharmaceutical industry (Dukes, 2006). For these reasons, there is a huge responsibility on the shoulders of plant taxonomists, who, unfortunately, often have to contend with a number of challenges including the intricate nature and complexity of plant life, and variability in their characteristics (Tilling, 1987), perceived tediousness of taxonomic practices along with obsolete tools for identification (Stagg and Donkin, 2013) and the attendant declining interest in botany (Drea, 2011), especially plant taxonomy (The Conservation, 2020). The number of botany specialists is reducing by the day. In the United States of America, the number of undergraduate degrees earned in botany is said to have decreased by 50% since the late 1980s (Bidwell, 2013), a trend that should never be taken lightly. The aforementioned challenges therefore formed the basis for conceptualising this study to make a contribution towards ameliorating the declining interest in plant taxonomy.
A taxonomic key is derived from a data matrix of a given number of ‘objects’, and it is usually possible to contrive a large number of different key formats for one set of objects such as plant species, but the keys will vary in their usability (Pankhurst, 1970). Dichotomous key is widely acknowledged as the most popular type of identification key (Sinh et al., 2017), and had been a clever means of organising taxonomic information before the age of computers (Godfray et al., 2007). The use of this key format is known to have contributed to increasing the quality and durability of knowledge of plant classification acquired in comparison to traditional teaching techniques (Andic et al., 2019) and an established method for teaching plant identification skills (Stagg and Donkin, 2013). However, a number of seemingly demoralising weaknesses are associated with dichotomous key format, including: being tedious to construct (Lobanov, 2003), having fixed point of entry and daunting path of navigation (Hagedorn et al., 2010), the problem of unanswerable couplet (Rambold and Martellos, 2010), being unusable for confirmation of suspected identity, and non-readily amenable to automation (Yin et al., 2016). So, invention of new key formats shall continue to be a welcome development in taxonomy. In providing a way out of the challenges enumerated earlier, the present study aimed at making the practice of plant taxonomy more attractive, less laborious and dreaded, and so, the objectives are to propose two new taxonomic key formats with highlights of their features, construction procedures and usage that should possibly make them desirable, either as alternative or complementary tools for plant identification.
Adoption of heuristic approach to addressing the weaknesses in dichotomous key format
The first step taken in actualising the objectives of this study was to align with the thoughts of Pankhurst (1970) on the two complementary problems in taxonomy, which are still valid till date. Firstly, given a set of objects (e.g., plants), examine their characteristics in order to find a classification, that is, group the objects into subsets (or taxa), and assign names to the subsets; and secondly, given a classification and an object, identify that object. In other words, given a list of the characteristics of named subsets which are known to exist, and an additional object, decide which subset the object belongs (that is, recognise it, or find its name). Accepting that the taxonomists’ diagnostic key was an important tool in the process of identification, the format and styles of the frequently used single-access diagnostic keys along with the challenges associated with their features, construction and application were critically examined (Walter and Winterton, 2007). In an effort to address some of these inadequacies, consideration was also given to selection criteria for construction of efficient diagnostic keys (Payne, 1981, 1988). Information obtained from the steps highlighted earlier were integrated into a thought to develop two alternative key formats, namely: the multi-level table and multi-layer circular diagnostic chart, each with far reaching desirable qualities in terms of design/features, construction, navigation efficiency and possibility of automation.
Data procurement for purpose of illustration
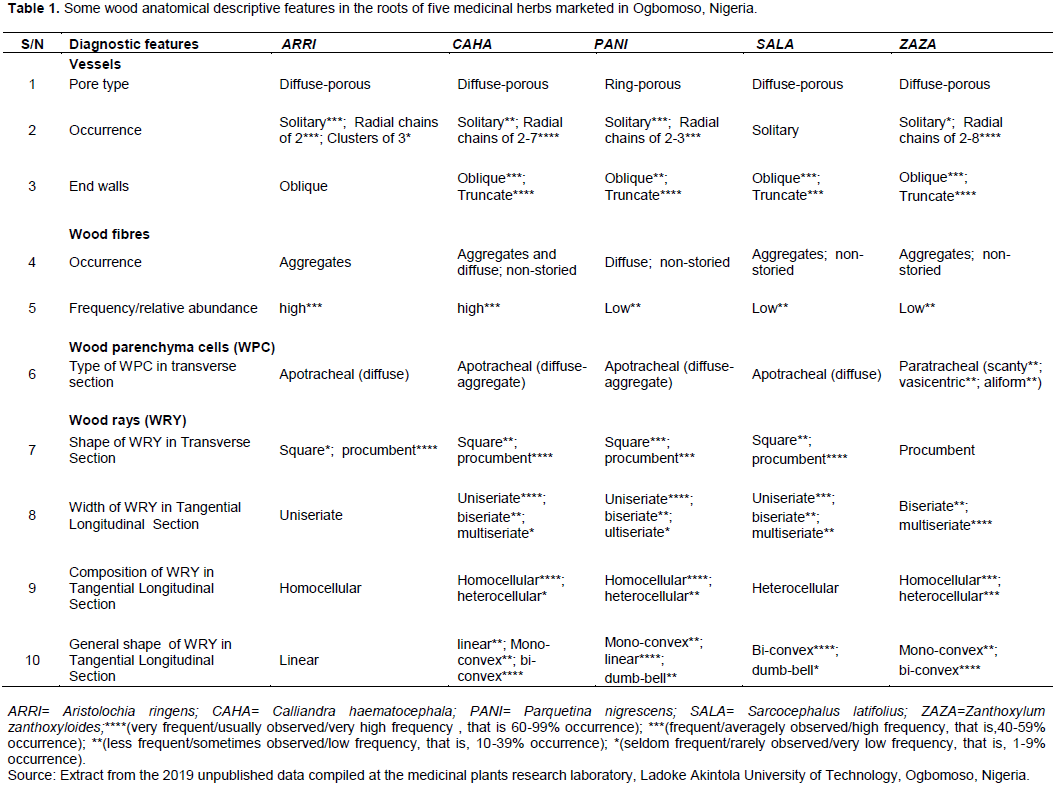
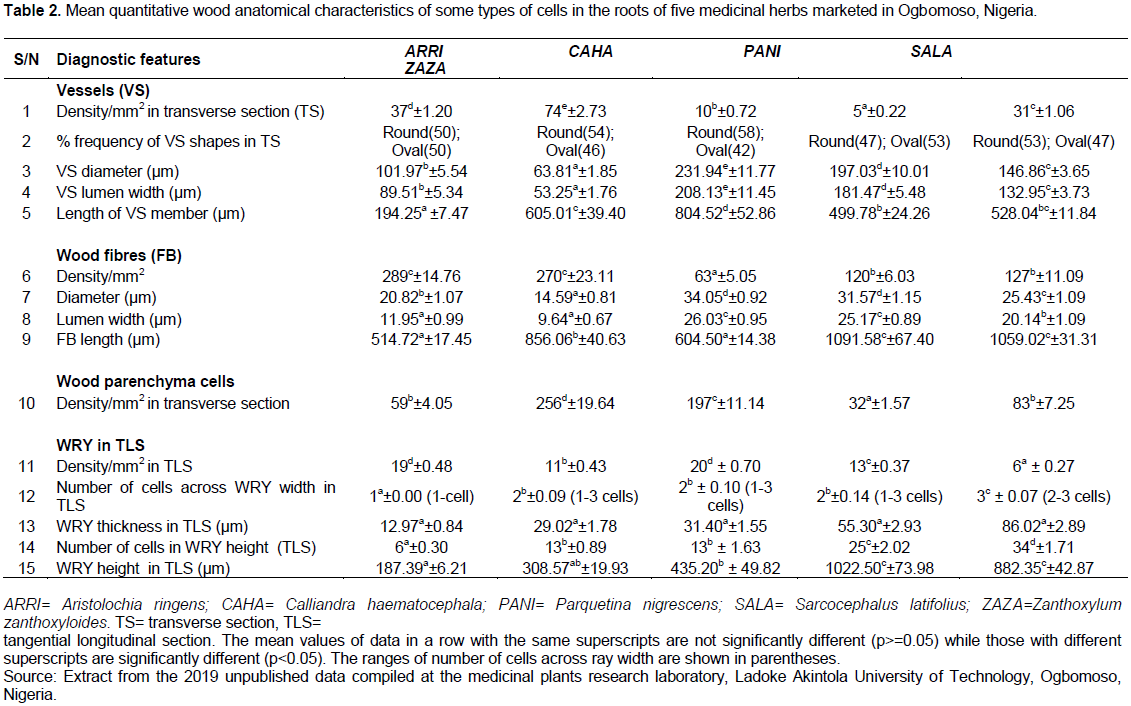
Wood anatomical data on five medicinal herbs marketed as plant roots in Ogbomoso township, south western Nigeria were sourced for the purpose of illustration from the 2019 compilation of unpublished results at the medicinal plants research laboratory in the Department of Pure and Applied Biology, Ladoke Akintola University of Technology, Ogbomoso, Nigeria. The species are Aristolochia ringens, Calliandra haematocephala, Parquetina nigrescens, Sarcocephalus latifolius and Zanthoxylum zanthoxyloides. The data items were obtained in accordance with the standard procedures: tissue sectioning/maceration (Schoch et al., 2004), staining, dehydration (Ogunkunle and Oladele, 2014), mounting (Arx et al., 2016), and microscopic observations (de Parnia and Miller, 1991), while the terminology and descriptions of observed features followed those of the International Association of Wood Anatomists (IAWA Committee, 1989). Twenty-five characters, consisting of ten qualitative (Table 1) and fifteen quantitative features (Table 2), all of which were diagnostic of the species were compiled. Staining was done in 1% alcoholic safranin, mounting was carried out in Canada balsam and observations made using Olympus biological microscope CH20i Model with binocular facility. Quantitative characters were considered diagnostic of the species only if the means of the replicated values were statistically significant at α = 5 following One-Way Analysis of Variance, and Duncan multiple range classification of the means (Landau and Everitt, 2004).
Conceptualisation of procedures for constructing and applying the two alternative key formats
For a given number of taxa with certain observable characters, the features as well as the procedures for constructing and navigating a multi-level table of identification on one hand, and the multi-layer circular diagnostic chart on the other hand were heuristically conceptualised as recursive or repetitive process of ‘divide and conquer’ algorithms (Hagedorn et al., 2010). Further to the achievement of the objectives of this study, the algorithm in each case was systematically executed, and is here, being proposed as a number of logical steps.
Design and statement of the features of multi-level table of identification
The multi-level table of identification was conceived as a diagnostic tool having features similar to the conventional table of results displaying characters and plant taxa in columns and rows. Unlike in a conventional table of results, the characters in the key are stated either as unit characters or combinations of two or more features observable in either a taxon or in clusters of taxa. Also unlike in the conventional table of results, the characters are arranged in tiers or levels: primary, secondary, tertiary, quaternary, quinary, senary, septenary, octonary, nonary denary, etc., representing first, second, third, …tenth level, respectively; indicating the levels of successive classification/compartmentalisation of the plant group using the characters. Characters of the first tier/level (that is, of the primary classification of the taxa) are listed in the first row on top of the table while the next row is used to display all the names of the taxa (that is, a cluster of plants) as defined by each character or characters in the first/primary level; characters of the second tier/level are listed in the third row of the table while the fourth row is used to display the clusters of taxa as defined by each character or characters in the second level, etc., thus rows of lists of characters alternate with rows of lists of taxa, and at the end of each successive level, the number of taxa in a cluster progressively reduces down the line.
Construction procedure of multi-level table of identification
The essential activities in building the multi-level table of identification consist of the following steps:
(1) constructing a conventional table of character comparison for the plant group under study, displaying characters in rows and the taxa in columns as in Tables 1 and 2;
(2) selecting one, or few characters as character combinations, which is/are considered as being of primary importance for classifying the group under study into few clusters of taxa that may be mutually or non-mutually exclusive, and stating those characters in the first row of the table; meanwhile, maximum number of clusters should be four to avoid the key being unwieldy;
(3) enumerating the taxa in each of the few clusters in the next row of the table as defined by the characters or character combinations in the previous row;
(4) considering the clusters of taxa obtained from primary level of classification, one at a time, selecting another character or character combination as being of secondary importance to further circumscribe the taxa in the primary cluster into smaller clusters (again, mutually-exclusive or not), and enumerating such taxa as a subset of that cluster in the next row;
(5) considering again, each cluster of taxa obtained from second tier of characters, selecting another, or few characters as being of tertiary importance to further circumscribe the taxa therein and enumerating such taxa as a subset of the secondary cluster in the next row;
(6) recursively selecting one or few characters (of quaternary, quinary, senary, etc., importance) for subsequent circumscription of the taxa as earlier described until every taxon in each cluster of taxa has been sorted/keyed out separately towards the bottom of their respective columns in the table;
(7) noting that if mutually exclusive clusters of taxa were possible and achievable throughout the recursive classifications of the group in steps ‘2’ to ‘6’, each taxon would be keyed out only once. However, if mutual exclusivity of the clusters was not achievable throughout the entire classifications, that is, if one or more non-mutually exclusive clusters were involved, at least one taxon would be keyed out more than once in the table.
Application of multi-level table of identification
In order to apply the multi- level table of identification, the following steps were formulated, adopted and are herein proposed:
(1) Evaluate the plant material based on the provisions of the first level of character combinations and decide which of the few clusters of taxa in the next row the plant belongs;
(2) Once a cluster of taxa has been selected in the row as the likely group in which the plant belongs, the few other clusters of taxa and the columns in which they fall should no longer be considered in subsequent steps;
(3) Re-evaluate the plant on the basis of the provisions of the next level of character combinations to decide which of the clusters of taxa in the next row the plant belongs; and if a decision is difficult or impossible because the plant features do not match the stated character combinations in the row, refrain from making a selection, but proceed to evaluate the unknown plant material on the basis of the characters in the next level;
(4) Repeat step 3, with the assurance that the features of the plant being identified align with those in the stated character combinations until a single taxon is achievable, which is taken as the identity of the unknown plant.
Design and statement of the features of multi-layer circular diagnostic chart
The multi-layer circular diagnostic chart was conceived as
consisting of two parts: the first part is a number of concentric circles, not drawn to scale, but partitioned by means of radial lines into a number of sectors, and the rings into compartments or ‘cells’; each sector representing a taxon in the key as indicated at the circumference; and each compartment is either assigned a number 1 or 2 or 3, etc., or is left void/empty as the case may be. The second part of the key is a list of characters or character combinations, assigned numerical values 1, 2, 3, etc., pertaining to the plant taxa in the circular diagram as appropriately indicated in the compartments.
Procedure for constructing a multi-layer circular diagnostic chart
The essential activities/steps in constructing the multi-layer circular diagnostic chart are as follows:
(1) A multi-level table of identification is first constructed as earlier described in steps ‘1’ to ‘7’;
(2) All the characters or character combinations in the table of identification obtained in step ‘1’ above are serially numbered 1, 2, 3, etc., from top to bottom, column after column, from the left to the right;
(3) A circle is drawn (not to scale) with a number of concentric rings not less than the number of levels/tiers of characters in the table of identification;
(4) The circle is divided into sectors equal to the number of taxa in the key, thereby partitioning the concentric rings into ‘cells’, boxes or compartments, and each sector is assigned a name of taxon at the circumference;
(5) The compartments in the circle are labeled using information in the appropriate table of identification by following a number of sub-steps:
(a) Consider the first taxon in the circular diagram, proceed to the first level of characters in the appropriate table, examine one column in the table after the other, and take note of every number (earlier assigned in step 2 above) attached to the character combinations 1, 2, 3, etc., that are contributors to the classification of the taxon being considered;
(b) Transfer those numbers attached to the relevant character combinations for the identification of the taxon into the compartments in the circle in reverse order, that is, the appropriate number at the first (topmost) level in the table is inserted in the last (innermost) compartment for the taxon; the next appropriate number at the next relevant level of characters down the table is inserted in the next upper compartment in the diagram and so on. Once the taxon is observed to have been keyed out in the table, subsequent transfer of numbers attached to character combinations for the taxon should be done in parentheses, indicating such characters to be of secondary importance, that is, although they are, or may be diagnostic of the taxon or a cluster of taxa, such characters need not be observable for taxa recognition to occur;
(c) Consider the next taxon in the diagram and repeat sub-steps ‘a’ and ‘b’;
(d) Remove any extra, entirely empty rings of compartments at the periphery of the circle, and the first part of the key would have been achieved;
(e) Compile, as an attachment to the circular chart, a list of all the characters/character combinations in the chronological order of numbering in the appropriate table, that is, 1, 2, 3, etc., thereby achieving the second part of the key.
Application of multi-layer circular diagnostic chart
In order to apply the multi-layer circular diagnostic chart for identification, the user enters the key at the centre and proceeds centrifugally (toward the circumference) by selecting the characters observable on/applicable to the unknown plant specimen at the successive rings of compartments. This process progressively narrows down on the choice of the possible identities (that is, sectors) of the unknown specimen until only one choice is achievable, which represents its identity.
Illustrative execution of the proposed procedures
The proposed steps for constructing and using the two new key formats developed from this study were executed using the wood anatomical data in Tables 1 and 2 to obtain two single-entry diagnostic keys usable for identifying five medicinal herbs sold as roots in Ogbomoso, Nigeria.
Tables 3 and 4 and Figures 1 and 2 are the results obtained following execution of the proposed procedures for constructing and using two alternative key formats. Construction of a multi-level table of identification (Tables 3 and 4) is a major step in making the multi-layer circular diagnostic chart (Figures 1 and 2), and both of these types of key are single-access devices. While Table 3 and Figure 1 were products of classifications of the five taxa into two mutually exclusive groups, Table 4 and Figure 2 are the results of classification of the taxa into three non-mutually exclusive groups.
Narrowing the lines of demarcation between the major components of taxonomy
In constructing a taxonomic key, an important suggestion by Radford et al. (1974) is to “identify all groups to be included in the key and prepare a description of each taxon”. This position has not changed till date. Morse (1971) explained that in preparing a key, one usually divides the initial group of taxa by a character couplet into two subgroups, each of which is independently divided into further subgroups, and so forth, until every taxon is distinguished from all others. Again, this procedure is valid till date, more so with a recent consenting publication (Hagedorn et al., 2010) regarding keys as ‘divide and conquer’ search algorithms that reduce the result set recursively until the remainder is small enough to be solved by direct comparison. These submissions point to the fact that although, identification is a separate activity in systematics, but in practice, it involves the other three major components of taxonomy namely classification, description and nomenclature (Radford et al., 1974). This scenario played out in the course of this study because the three activities were brought to bear in developing two new key formats for the purpose of identification.
Examining beliefs and opinions on identification keys
One implication of adopting the above-stated suggestion by Radford et al. (1974) is that the author of a key should or will not, ordinarily require same key to identify any of the taxa included in it. Ab-initio, he identified all the taxa and created the key. If one views this scenario on the surface, along with the general belief that the use of identification keys requires intensive training and experience, which only few individuals do have (Waldchen et al., 2018), one will agree with Lobanov (2003) that “keys are compiled by those who do not need them for those who cannot use them”. However, on a closer examination of the two pillars on which Lobanov’s conjecture reclines, one would tend to disagree with him. Firstly, a taxonomist is not expected to have worked on all plant groups, nor is he obliged to keep in mind separately the names and diagnostic features of those taxa included in all the keys he has authored. Therefore, as a specialist, he not only uses keys created by his colleagues who are specialists in various other plant groups, those written by him are also potential tools for him to carry out identification of the taxa afterwards. Considering the second leg of the argument, it is to be understood that writers and users of keys do not necessarily occupy mutually exclusive positions, so the question of certain persons outside of a clique not being able to use identification keys ought not to be overstressed in the first place.
One valid deduction from Lobanov’s hypothesis on identification keys is that there is ‘plentiful harvest, but few workers to gather it in’, and this position is explainable as follows: Identification is the basic prerequisite to understanding biodiversity and ecology (Randler, 2008), and is indispensable in many facets of human life. However, species identification is perceived by many practitioners as onerous task, being comparable with the learning of new words of a new language, while others believe that the act is much more difficult and complex (Randler, 2008). Arising from this perception, more and more students and researchers are showing reduced interest in taxonomy. True, there are many non-botany specialists who desire correct identification of plants but are constrained by lack the technical know-how. The up-coming botany specialists who ought to be of assistance to the former category of people in this regard are not better-off either. Furthermore, the current rates of species lost to extinction (IUCN, 2017) necessitate concerted attempts to protect and conserve biodiversity. Species conservation, however, requires species identification skills, but there seems to be perpetual shrink in the number of those interested in this mission (Woodland, 2007).
Comparing the structural and functionality attributes of dichotomous key, multi-level table of identification and circular diagnostic chart
Although enumeration of the features of dichotomous keys is not included in the core objectives of this study, a brief highlight of the challenges associated with the construction and use of this important key format will pave the way for a hands-on approach to comparing the features of the newly proposed key formats with those of the dichotomous format, a single-access identification tool with which key users are most familiar (Sinh et al., 2017). In a dichotomous key, as well as in each of the two newly developed keys in this study, there is only one point of entry, so that there is a single path to be followed by the user. This is a property shared by all single- access key formats, and is accompanied by the problem of ‘unanswerable couplet’, that is, a user may get stuck and identification will be impossible if a choice cannot be decided at any point (Hagedorn et al., 2010). Also, of concern to users of single access keys are the issues of ‘dead ends’, and the ‘momentary distractions’ that can cause a user to forget his or her position in a key (Walter and Winterton, 2007). These situations can arise when a character cannot be observed or adequately scored (e.g. when the feature is in its developmental stage or is season-based, and hence not visible in the specimen) or because the options are not stated clearly enough in the key. The magnitude of the frustration that may set in due to these problems can be intolerable, especially to novice taxonomy students. While the dichotomous key format is notoriously prone to these challenges, such difficulties can be more tolerable with the application of the multi-level table and circular diagnostic chart being proposed since it is much easier to retrace one’s steps in case a wrong choice has been made.
It is the belief in certain quarters that construction and use of dichotomous keys are daunting tasks for many students (Jacquemart et al., 2016), and that the format is difficult to automate, if at all amenable to conventional programming techniques (Yin et al., 2016). In contrast, both the construction and navigation procedures of the newly developed single access key formats, that is, multi-level table and circular diagnostic chart have clear-cut algorithms, which can be followed by key makers and users with relative ease. Additionally, these algorithms can be coded using the desired programming languages; so automation of these activities should not be an intractable problem.
While both the paper-based and computerised dichotomous keys (Tofilski, 2018) are not readily usable for confirmation of suspected identity of a plant, this exercise is manually practicable and electronically achievable using the two newly created single access key formats in this study. If for an unknown plant specimen, one of the taxa included in a key is suspected by a user as its identity, the procedure to confirm or otherwise is first locate the position of the suspected taxon in a key and then work on the key along the established route of identifying the taxon, paying particular attention to only those statements/questions regarding the suspected taxon name, and ensuring that all such (not most) statements are in agreement with the observable features of the specimen in the hand. For the purpose of illustration, if a user in applying the key in Table 3 suspects the identity of a plant to be C. haematocephala, confirmation is done by first locating the positions at which the suspected name has been successively keyed out in the column on the left and then the specimen is evaluated based on those characters, that is, the first, second and fourth level of characters where the taxon name occurs. Similarly, if the same taxon is suspected using the key in Figure 1, the specimen is evaluated based on characters 1, 2 and 4 only. So, if given a key, and the assurance that a suspected taxon is included in that key, plant identity confirmation can be explored as a means of assessing leaners’ extent of familiarity with the vegetation around them. The learners will not only find the exercise pleasing and refreshing, but also inspiring, much like a game.
Arresting the declining interest in botanical knowledge
Stagg and Donkin (2013) believed that the demise of botanical interest was due to the way botany was taught, if it was taught at all. In order to curtail this undesirable development, appealing plant identification resources are needed, making botany relevant to people’s lives is necessary, and correct use of new teaching aids is important (Tilling, 1987). Each of the two newly designed identification key formats in this study has provided answers to these calls. With the likes of multi-level table of identification (Tables 3 and 4) and circular diagnostic chart (Figures 1 and 2) in place, taxonomic key construction and use for identification or identity confirmation turn out to be favourite pastime for specialists and novices alike. In lieu of dichotomous keys, the circular diagnostic chart has additional merit of being adaptable (possibly with enhancements in form of multiple attractive colours) for use at the primary school level, or as braille, with or without sound effects, for use by visually challenged persons (Andic et al., 2019).
CONCLUSIONS AND RECOMMENDATIONS
In this study, two new taxonomic key formats have been designed, illustrated and proposed for use in plant taxonomy. They are namely: multi-level table of identification and multi-layer circular diagnostic chart, both of which are single access devices. Using these key formats, the trio activities of key construction, plant identification, and plant identity confirmation are made possible through robust algorithms. Since each of these algorithms is in conformity with the principal features of a good/executable computer algorithm (that is, being deterministic, general, finite, and with capacity to act on at least one input to produce at least one output), it is believed that these alternative key formats should be programmable. Going by their features and functionality attributes, the two new key formats proposed in this paper are recommended as useful templates upon which reliable plant diagnostic tools can be based. This paper has also contributed wood anatomy-based diagnostic keys usable for authenticating five medicinal herbs marketed as plant roots in Ogbomoso, Nigeria.
The author has not declared any conflict of interests.
The inspiration to create alternative key formats for plant identification was received from the emboldening words of late Prof. Felix Oladele. The author thanks Prof. Aderemi Okeyinka of Department of Computer Science, Landmark University, Omu-Aran, Nigeria for the helpful comments and pieces of advice on the draft manuscript, his students at the Medicinal Plants Research Laboratory, Ladoke Akintola University of Technology, Ogbomoso, Nigeria, and Mrs. Jennifer Ideh and Mr. Gideon Olaniran for assisting to collate the data used for illustration.
REFERENCES
|
Amante VD, Norton GA (2003). Developing interactive diagnostic support tools for tropical root crops. Centre for Biological Information Technology, The University of Queensland, Brisbane Qld 4072, Australia.
|
|
|
|
Andic B, Cvijeticanin S, Maricic M, Stesevic D (2019). The contribution of dichotomous keys to the quality of biological knowledge of eighth grade students. Journal of Biological Education 53(3):310-326.
Crossref
|
|
|
|
|
Arx GV, Crivellaro A, Prendin AL, Cufer K, Carrer, M (2016). Quantitative wood anatomy- practical guidelines. Frontiers in Plant Science 7:781.
Crossref
|
|
|
|
|
Bidwell A (2013). The academic decline: how to train the next generation of botanists. Available at:
View.
|
|
|
|
|
Bock JH, Norris DO (2016). Forensic plant taxonomy. In: Forensic plant science, Bock JH and Norris DO, Academic Press. pp. 95-101.
Crossref
|
|
|
|
|
De Parnia NE, Miller RB (1991). Adapting the IAWA list of microscopic features for hardwood identification to DELTA. IAWA Journal 12(1):34-50.
Crossref
|
|
|
|
|
Drea S (2011). The end of the botany degree in the UK. Bioscience Education 17(1):1-7.
Crossref
|
|
|
|
|
Dukes G (2006). The law and ethics of the pharmaceutical industry. Elsevier Science Publisher, B.V. The Netherlands.
|
|
|
|
|
Godfray HCJ, Clark BR, Kitching IJ, Mayo SJ, Scoble MJ (2007). The web and the structure of taxonomy. Systematic Biology 56(6):943-955.
Crossref
|
|
|
|
|
Hagedorn G, Rambold G, Martellos S (2010). Types of identification keys. In: Tools for identifying biodiversity; progress and problems. Nimis PL and Vignes LR (eds.), EUT. pp. 59-64.
|
|
|
|
|
Hopkins B, Stanfield DP (1966). A field key to the savanna trees in Nigeria. Ibadan University press, Ibadan.
|
|
|
|
|
IAWA Committee (1989). International Association of Wood Anatomists (IAWA) list of microscopic features for hardwood identification. IAWA Bulletin 10:219-332.
Crossref
|
|
|
|
|
IUCN (2017). The IUCN red list of threatened species, International Union for Conservation of Nature.
View
|
|
|
|
|
Jacquemart AL, Lhoir P, Binard F, Descamps C (2016). An interactive multimedia dichotomous key for teaching plant identification. Journal of Biological Education 50(4):442-451.
Crossref
|
|
|
|
|
Javatpoint (2018). DBMS keys, database management systems. Available at:
View
|
|
|
|
|
Jones A, Reed R, Weyers J (1998). Practical skills in biology (2nd ed.). Addison Wesley Longman limited. London.
|
|
|
|
|
Landau S, Everitt BS (2004). A handbook of statistical analyses using SPSS. Chapman and Hall/CRC Press Company, Washington, D. C. Available at:
View
|
|
|
|
|
Lobanov AL (2003). Keys to beetles and biological diagnostics.
|
|
|
|
|
Lowe J, Stanfield DP (1974). The flora of Nigeria. Sedges (family Cyperceae). Ibadan University press, Ibadan.
|
|
|
|
|
Morse LE (1971). Specimen identification and key construction with time -sharing computers. Taxon 29:269-282.
Crossref
|
|
|
|
|
Ogunkunle ATJ, Oladele FA (2014). Contributions to classification of some Nigerian species of Ficus L. (Moraceae) by using wood anatomical characteristics. Journal of Plant Biology Research 3(1):23-36.
|
|
|
|
|
Pankhurst RJ (1970). A computer program for generating diagnostic keys. The Computer Journal 13(2):145-151.
Crossref
|
|
|
|
|
Panter KE, Welch KD, Gardner DR (2014). Poisonous plants: biomarkers for diagnosis. In: Biomarkers in Toxicology, Gupta RC (ed.). Elsevier, Oxford, UK. pp. 563-589.
Crossref
|
|
|
|
|
Payne RW (1981). Selection criteria for the construction of efficient diagnostic keys. Journal of Statistical Planning and Inference 5(1):27-36.
Crossref
|
|
|
|
|
Payne RW (1988). Identification keys, diagnostic tables and expert systems. In: Edwards D and Raun NE (eds.). Compstat, Physica-Verlag Heidelberg.
Crossref
|
|
|
|
|
Payne RW, Walton E, Barnett JA (1974). A new way of representing diagnostic keys. Journal of General Microbiology 83(2):413-414.
Crossref
|
|
|
|
|
Payne RW, Preece DA (1980). Identification keys and diagnostic tables: A review. Journal of the Royal Statistical Society: Series A (General) 143(3):253-292.
Crossref
|
|
|
|
|
Price PD (1967). Two Types of Taxonomy: A Huichol Ethnobotanical Example. Anthropological Linguistics 9(7):1-28.
|
|
|
|
|
Radford AE, Dickison WS, Massey JR, Bell CR (1974). Vascular Plant Systematics. Harper and Row. New York.
|
|
|
|
|
Randler C (2008). Teaching species identification- a prerequisite for learning biodiversity and understanding ecology. Eurasia Journal of Mathematics, Science and Technology Education 4(3):223-231.
Crossref
|
|
|
|
|
Schoch W, Heller I, Schweingruber FH, Kienast F (2004). Wood anatomy of central European Species. Swiss Federal Institute for Forest.
|
|
|
|
|
Sinh NV, Wiemers M, Seettele J (2017). Proposal for an index to evaluate dichotomous keys. Zookeys 685:83-89.
Crossref
|
|
|
|
|
Stagg BC, Donkin MC (2013). Teaching botanical identification to adults: experience of the UK participatory science project open air laboratories. Journal of Biological Education 47(2):104-110.
Crossref
|
|
|
|
|
The Conservation (2020). Botany may be dying, but somehow the plants survive. The Conservation Africa Inc. Available at:
View.
|
|
|
|
|
Tilling SM (1987). Education and taxonomy: the role of the field studies council and AIDGAP. Biological Journal of the Linnaean Society 32(1):87-96.
Crossref
|
|
|
|
|
Tofilski A (2018). DKey software for editing and browsing dichotomous keys. Zookeys 735:131-140.
Crossref
|
|
|
|
|
Rickett HW (1958). So what is a Taxon? Taxon 7(2):37-38.
Crossref
|
|
|
|
|
Upton R, Romm A (2010). Guidelines for herbal medicine use. In: Botanical medicine for women's health, Romm A, Hardy ML, Mills S (eds.). Churchill Livingstone, pp. 75-96.
Crossref
|
|
|
|
|
Walter DE, Winterton S (2007). Keys and the crisis in taxonomy: extinction or reinvention? Annual Review of Entomology 52:193-208.
Crossref
|
|
|
|
|
Waldchen J, Rzanny M, Seeland M, Mader P (2018). Automated plant species identification- trends and future directions. PLoS Computational Biology 14(4):e1005993.
Crossref
|
|
|
|
|
Woodland DW (2007). Are botanists becoming the dinosaurs of biology in the 21st Century?. South African Journal of Botany 73(3):343-346.
Crossref
|
|
|
|
|
Yin S, Lin X, Liu L, Wei S (2016). Exploiting parallelism of imperfect nested loops on course-grained reconfigurable architectures. IEEE Transactions on Parallel and Distributed Systems 27:3199-3213.
Crossref
|
|