Full Length Research Paper
ABSTRACT
Groundnut root rot is a complex disease affecting the crop production worldwide. The objectives of this study were to: (1) Assess the occurrence and distribution of groundnut root rot complex in four main groundnut-growing areas of eastern Ethiopia; (2) Identify the major pathogens associated with groundnut root rot complex; and (3) Determine the role of pathogens singly and in combination on root rot severity of groundnut under greenhouse conditions. A total of 240 groundnut fields were surveyed over two seasons, and 75% of the fields were infected with root rot complex. Mean percent disease prevalence and incidence were highest in Babile, followed by Gursum, while Darolebu had the lowest. Fusarium oxysporum (Fo) was the most common fungus isolated from infected groundnut roots, followed by Fusarium solani (Fs) and Rhizoctonia bataticola (Rb). Other fungi less frequently isolated included Sclerotium rolfsii (Sr) and Rhizoctonia solani (Rs). The pathogenicity test showed that all isolated fungi were pathogenic to groundnut, with Fs being the most virulent in causing root rot, followed by Rb and Fo, while Sr and Rs were less pathogenic. Disease severity was higher on plants co-inoculated with Fo, Fs and Rb than with either alone. The Fs, Fo and Rb were found to be the major causal fungi associated with groundnut root rot complex in eastern Ethiopia, with Fs being the most important and destructive pathogen in the complex and acted as synergistic pathogen with Fo and Rb; thus, breeding for resistance and management strategies should focus on these pathogens.
Key words: Fusarium oxysporum, Fusarium solani, incidence, pathogenicity, pathogen interaction, prevalence, Rhizoctonia bataticola, severity.
INTRODUCTION
Groundnut (Arachis hypogaea L.), also known as peanut and belongs to the family Fabaceae (Nordern et al., 1982; Hassuba et al., 2016), is an important food and oilseed crop grown worldwide, with an annual production of 53.6 million tons (t) from an area of 31.6 million hectares (ha) in 2020 season (FAOSTAT, 2022). Africa and Asia account for about 95% of worldwide groundnut areas where it is grown under rainfed condition with little agricultural inputs by resource-poor farmers (Janila et al., 2013). In Ethiopia, groundnut is the second most important oilseed crop next to sesame, and largely produced in the eastern parts of the country (Amare et al., 2017). Its production was estimated at 205,069 tons from an area of about 113,515 ha with productivity of 1.8 t ha-1 in 2020 cropping season (FAOSTAT, 2022). Groundnut is an important source of cash income for farming families and generates foreign currencies for the country (Gezahagn, 2013). The crop contributes to nutrition of farmers through utilization of high energy- and protein-rich groundnut seeds, and also provides nutritious fodder/haulms to their livestock (Janila et al., 2013). Groundnut also has an important role in sustaining agricultural systems by improving soil fertility through biological nitrogen fixation, and crop yield thereby reducing reliance on inorganic fertilization (Ajeigbe et al., 2014).
Despite its considerable importance, the average yield of groundnut in Ethiopia is much lower than other groundnut-growing regions, such as China, Egypt, Indonesia and USA (FAOSTAT, 2022). This low yield is mainly due to several biotic and abiotic factors, which include critical moisture stress, lack of improved cultivars, low soil fertility content and poor crop management practices; and plant diseases infecting both the above and underground parts of the plants, and aflatoxin contamination which infect both the crop yield and quality in the field and at different levels from harvest to market in the value chain (Chala et al., 2014; Meresa and Tsehaye, 2020). Early leaf spot (caused by Cercospora arachidicola) and late leaf spot (caused by Cercosporidium personatum) and rust (caused by Puccinia arachidis) are the major foliar diseases of groundnut in eastern Ethiopia. Root rot complex is a common and growing problem for groundnut production worldwide. Several different soil-borne pathogens, including Fusarium solani, Fusarium oxysporum, Rhizoctonia solani, Sclerotium rolfsii, and Rhizoctonia bataticola (synonym = Macrophomina phaseolina) have been reported to be involved with groundnut root rot complex from various regions of the world (Atta-Alla et al., 2004; Mahmoud et al., 2006; Sharma et al., 2012; Mahmoud, 2015; Bodah, 2017).
Groundnut root rot complex has received limited attention compared to foliar plant diseases, with most studies tending to focus on a single pathogen. The disease caused by two or more pathogens often is more severe and damaging than that induced by one pathogen because of their synergistic effects (Lamichhane and Venturi, 2015). This pathogen interaction influences disease intensity and plant growth (Porto et al., 2019; Sumbul and Mahmood, 2020). Depending on the pathogen(s) associated, common disease symptoms might include any combination of the following: damping-off, poor seedling establishment in field, irregular growth with poor stand, chlorosis, wilting and yellowing of leaves, stunted plant growth, death of severely affected plants, reduced yield, quality and loss of crop. Affected plant roots are also reduced in sizes, discolored, and display different stages of decay and defoliation (de Jensen, 2000; Rajamohan and Balabaskar, 2016). Root rots are greatly influenced by environmental condition, and is favored by low-and -high soil-moisture, drought stress, optimum temperatures for pathogens development, soil compactions, continuous monocropping and other different factors that contribute stresses to plants (Rojo et al., 2007; Rajamohan and Balabaskar, 2016; Bodah, 2017).
In recent years, groundnut root rot complex has become a major constraint to groundnut production in eastern Ethiopia, where the crop is intensively cultivated in the country. Its outbreaks negatively affected the livelihood of several small-scale farmers who totally depend on this crop as a major source of food and cash income (Tarekegn et al., 2007). Fusarium species as major and occasional association of R. bataticola and S. rolfsii with root rot affected groundnut was reported (Tarekegn et al., 2007). However, this fact was based on limited research data conducted before a decade and was insufficient to develop an integrated management strategy for eastern Ethiopia groundnut root rot complex. Efforts in groundnut breeding for root rot resistance will be strengthen if the composition and relative importance of involved pathogens are well documented. Furthermore, an understanding of the interaction between the pathogens associated in the root rot complex is necessary to develop effective management strategies.
Therefore, the specific objectives of the present study were to: (1) assess the occurrence and distribution of groundnut root rot complex in main groundnut-growing areas of eastern Ethiopia; (2) identify the major pathogens associated with the groundnut root rot complex; and (3) determine the role of pathogens singly and in combination on root rot severity of groundnut under greenhouse condition.
MATERIALS AND METHODS
Description of field survey areas
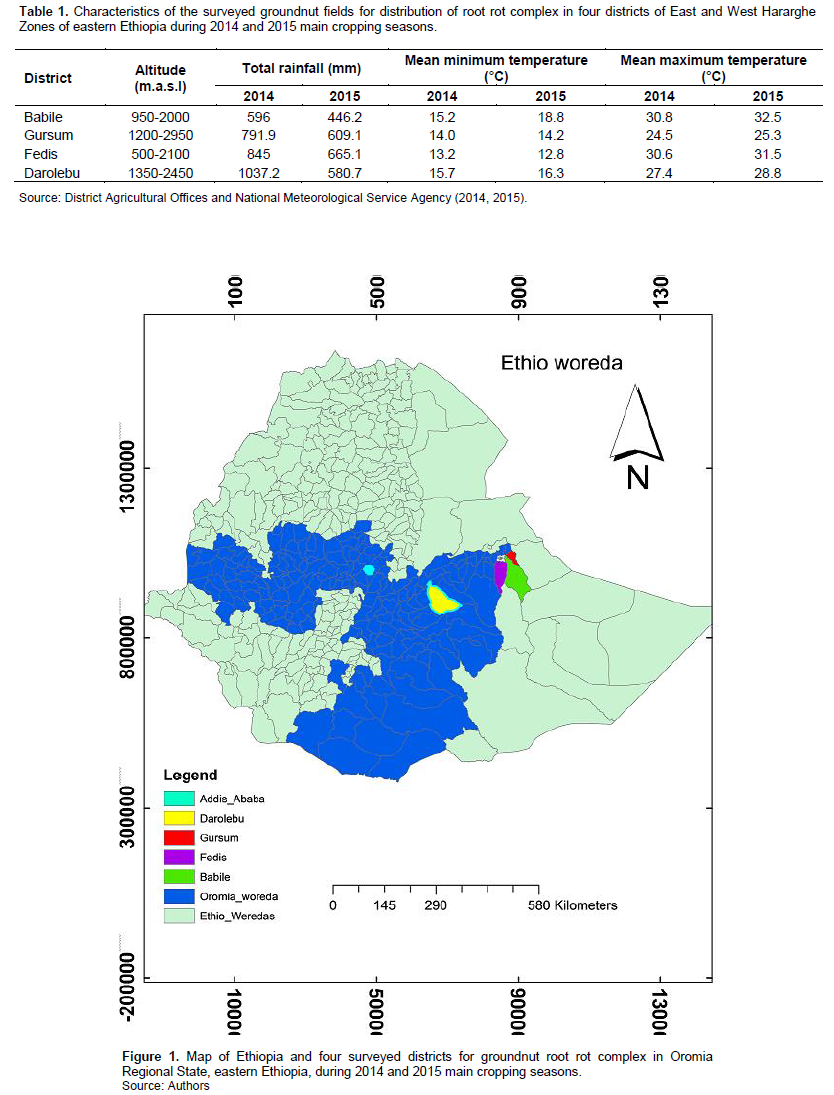
Two-year surveys were carried out in four main groundnut-growing areas of eastern Ethiopia during the 2014 and 2015 main cropping seasons. The four districts covered in the field surveys are found in two Administrative zones of Oromia Regional State: Babille, Fedis and Gursum districts are found in the East Hararghe zone, while Darolebu is found in the West Hararghe zone. These areas were selected because they are the main areas of groundnut cultivation in eastern Ethiopia as well in the country (CSA, 2017). East Hararghe zone is geographically located between 7°30’-9°45’N latitude and 41°10’-42°50’E longitude, with an elevation ranging from 500 to 3040 m above sea level (m.a.s.l.) (Tadesse et al., 2014), whereas West Hararghe is located between 7°32’-9°47’N latitude and 41°24’-43°48’E longitude, with an elevation ranging from 500 to 3500 m.a.s.l. (Kassa et al., 2013). The characteristic features of the areas surveyed and their geographical locations are depicted in Table 1 and Figure 1.
Field sampling and groundnut root rot complex disease assessment
A total of 240 groundnut fields (30 fields per district in each season) were surveyed over the 2014 and 2015 main cropping seasons. Based on the importance of the crop and root rot problems, 3 representative farmers’ administrations (FAs) were selected from each district. In each FA, 10 farmers’ groundnut fields selected at random were surveyed for detailed assessment of root rot complex. During both year field surveys, groundnut fields were sampled randomly at intervals of about 3 to 10 km along the main and accessible rural roads and distance between the fields depended on the relative importance of groundnut cultivation in every district. From each field, five quadrats (1.5 m × 1.5 m) were randomly sampled for groundnut root rot complex disease assessment by making a diagonal move in the field. Disease prevalence was recorded as the percentage of root rot affected groundnut fields, whereas disease incidence was assessed as the percentage of groundnut plants showing typical root rot symptoms in each quadrat, and means of 5 quadrats were calculated for each field in each district. The following formulae were used to calculate the percentage of disease prevalence and disease incidence:
From each field, three to four randomly selected diseased plants were collected for isolations and identification of associated pathogen(s) in the laboratory. Sampling of diseased plants was done by uprooting the whole plant from the soil with the help of spade. Effort was made to take out the entire plant root system by digging the roots carefully. After excising the aerial portion, the roots were placed in polyethylene bags and labeled properly. All labeled bags were tied and stored in ice-cool-box to prevent from desiccation and other contamination, and then brought to the Haramaya University Plant Pathology Laboratory for processing and analyses. The root rot samples were kept at 4°C in a refrigerator until fungal isolations were performed.
Pathogen isolation and identification
Symptomatic plant root samples were carefully washed under
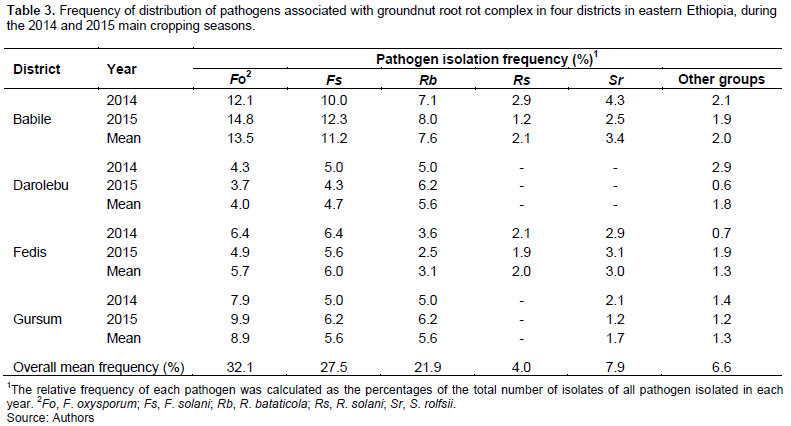
running tap water to any attached organic debris and soil particles, and then cut into about 1 cm portions using a sterile scalpel. These root pieces were surface-sterilized in 3% sodium hypochlorite (NaOCl) solution for 3 min, rinsed 3 times in successive changes of sterile distilled water (SDW) to remove the residue of sodium hypochlorite, and then dried between sterile filter paper under a laminar flow hood. These dried root segments (5 pieces per plate) were then aseptically placed onto potato dextrose agar (PDA) medium amended with chloramphenicol (50 mg L-1) in 90 mm Petri-dishes, incubated at 25±2°C for 3 to 7 days, and observed daily for any fungal growth. Fungal colonies emerging from plated root pieces were then individually subcultured onto new PDA plates, and purified with hyphal tip or single-spore transfer techniques. The organisms/isolates associated with the infected root rots were observed under the compound microscope. Fungi were then identified based on their cultural and morphological characteristics using laboratory manuals for identification keys (Burnett and Hunter, 2003). Then pure cultures of Fusarium spp. were further transferred onto carnation leaf agar (CLA) and identified to species levels (Nelson et al., 1983; Leslie and Summerell, 2006). Accordingly, five pathogenic fungal species were identified (Table 3). The relative frequency of each fungal pathogen was calculated as the percentage of the total number of isolates of all species obtained in each cropping season. Pure cultures of selected fungal isolates were maintained by transferring onto PDA slants in test tubes and were kept at 4°C in a refrigerator for further studies (Chopada et al., 2015).
Pathogenicity test
To determine the pathogenicity of fungi recovered from symptomatic groundnut roots, pathogenicity tests were conducted using sorghum grain inoculum techniques with three randomly taken isolates of each of five fungal species recovered from infected groundnut roots in this study. Briefly, sorghum grains were water-soaked overnight, air-dried under room temperatures in a laminar flow hood, and then placed in 250 mL conical flasks. The mouth of every flask was plugged with a cotton wool, wrapped in aluminium foils, and autoclaved (at 121°C, 15 psi for 45 min) for two consecutive days. After cooling, the sorghum grains in the flasks were inoculated with three 5 mm mycelial plugs from actively growing 7-day old cultures of each of the five fungal isolates (used as treatments) grown on PDA medium and incubated at 25±2°C for 15 days. All these flasks were shaken at alternate days for uniform colonization of the sorghum grains. Pathogen inoculum thus produced was used to infest steam-sterilized pot mixture of field soil, sand and farm yard manures in a ratio of 2:1:1 v/v. The soil infestation was done by mixing inoculum into the soil surface of each plastic pot (14 cm diameter × 18 cm height) at the rate of 5% w/w (Atta-Alla et al., 2004). The same amount of sterilized sorghum grains was added to other plastic pots to serve as a control check. Pathogen infested pots were watered and kept for 7 days before planting groundnut seeds, variety Oldhale, to ensure pathogen development (Mahmoud et al., 2013).
Apparently healthy looking seeds of local groundnut variety ‘Oldhale’, which is commonly produced by the farmers and susceptible to root rots (Tarekegn et al., 2007), were surface sterilized (Atta-Alla et al., 2004), and planted in pots filled with pathogen infested soil-mixture and the control at the rate of five seeds per pot. After emergence, two seedlings were removed to maintain three plants per pot. The pots were maintained in greenhouse at temperatures varying from 21 to 30°C under natural light condition. The treatments with the five fungal isolates in the infested soils in pots in the greenhouse were arranged in a completely randomized design with three replicates of each isolate, and the experiment was repeated once. The pots were watered as needed, and plants in each pot were assessed for root rot symptom development. Observations were also made frequently for the appearance and development of root rot symptoms.
Then eight weeks after planting, groundnut plants were removed from each pot, washed and rated for disease severity using a 0 to 5 root rot disease scale (Naseri, 2008) based on the percentage of plant root tissue with discoloration, lesion, or rots: 0 = no visible root symptoms; 1 = light discoloration or 1-10% covered with lesion; 2 = heavy discoloration, 10-25% covered with lesion, or light rot; 3 = 25-50% covered with lesion or rot; 4 = 50-75% covered with lesion or rot; and 5 = 75-100% rot and plants dead. The disease severity scores were then converted into severity index (SI) for analysis using the following formula (Wheeler, 1969):
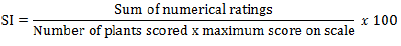
Each tested species of fungi was considered to be pathogenic when its inoculation to groundnut seedlings under greenhouse resulted in at least one diseased plant with typical root rot symptoms. To fulfill Koch’s postulates, fifteen randomly taken root pieces from the three replicated pots of every pathogen treatment were surface-disinfected (in 3% sodium hypochlorite solution for 3 min), rinsed 3 times in sterile distilled water, blotted dry, and then plated on PDA to check for the occurrence of the tested fungal species in root rot infected tissues. The plates were incubated at 25±2°C for 3 to 7 days, and their cultural and morphological features were recorded and compared with the original fungal isolates of each pathogen.
Pathogen interaction
Pathogen interaction experiment was carried out in the greenhouse to test the synergistic or antagonistic effect of the three commonly isolated fungal species on groundnut using sorghum grain inoculum technique as described above in the pathogenicity tests. The following fungal pathogens and their combinations were used in this study: (1) F. oxysporum (#Fo-20); (2) F. solani (#Fs-15); (3) R. bataticola (#Rb-10); (4) F. oxysporum (Fo) + F. solani (Fs); (5) F. oxysporum (Fo) + R. bataticola (Rb); (6) F. solani (Fs) + R. bataticola (Rb); (7) F. oxysporum (Fo) + F. solani (Fs) + R. bataticola (Rb); and (8) Control (sterile sorghum grains only). Seeds of a local groundnut variety ‘Oldhale’, which is susceptible to different root rot pathogens, were surface-sterilized (Atta-Alla et al., 2004), and planted in plastic pots filled with pathogen infested soil mixture as described earlier. The pots containing the treatments were arranged in the greenhouse in a completely randomized design with three replications, and the trial was repeated once. Fifteen randomly taken root pieces from the three replicated pots of every pathogen treatment were surface-disinfected (in 3% sodium hypochlorite solution for 3 min), rinsed 3 times in changed sterile distilled water, blotted dry, and plated into PDA to check, using compound microscope, for the occurrence of tested fungi in infected root tissues.
Data analysis
The disease prevalence and incidence data obtained from groundnut field surveys were analyzed with descriptive statistics (Gomez and Gomez, 1984). The disease data were statistically analyzed with the SAS software system (SAS Institute Inc., Cary, 2004). Homogeneity of variance between repeated trials was tested with Levene’s test. The data of repeated trials were combined and analyzed with GLM procedures, when the variance error between repeated trials was homogeneous. Mean separation was performed with Fisher’s protected least significance (LSD) test at 5% probability levels (Gomez and Gomez, 1984).
RESULTS
Occurrence and distribution of groundnut root rot complex
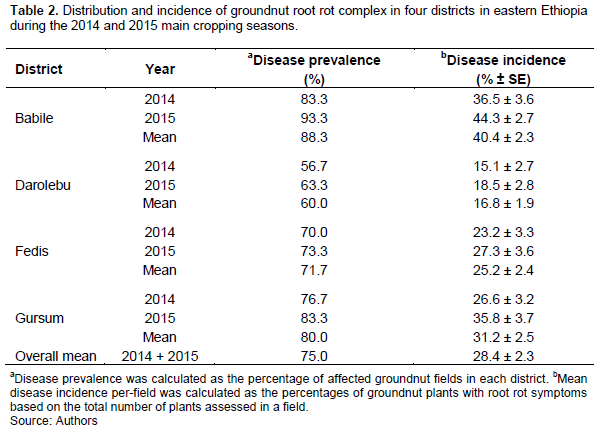
Groundnut plants with root rot symptoms were found in 75% of the fields inspected (Table 2). Groundnut in these fields displayed a number of disease symptoms, which could be assigned to the disorders typical for the root rot complex (Porch et al., 2014). The most prominent disease symptoms observed in these fields included yellowing and severe stunting of plants; discoloration and collapsing of outer leaves, which had discolored, rotted, and reduced root systems. The results indicate that disease prevalence and incidence symptoms differed among the four districts and between the two main cropping seasons. The highest (88.3%) mean disease prevalence value was computed for the Babile district, followed by Gursum (80.0%), and Fedis (71.7%), whereas Darolebu fields showed the lowest (60.0%) mean disease prevalence. The same trend was also detected with mean disease incidence, which ranged from 16.8% in Darolebu to 40.4% in Babile fields. Disease prevalence and incidence in 2015 were greater by 3-10 and 3-9%, respectively, than in 2014 cropping season (Table 2).
Pathogens associated with groundnut root rot complex
A total of 302 fungal isolates: 140 isolates in 2014 and 162 isolates in 2015 main cropping seasons were recovered from diseased groundnut root samples collected in four districts of East and West Hararghe zones, eastern Ethiopia. Among these, five fungal species were identified based on their cultural and morphological characteristics (Table 3 and Figure 2). The most (32.1%) common or frequent pathogen isolated over the two main cropping seasons was F. oxysporum, followed by F. solani (27.5%), R. bataticola (21.9%), S. rolfsii (7.9%), and R. solani (4.0%). Except S. rolfsii and R. solani, all the three isolated fungal species were found in all four districts that were surveyed. The S. rolfsii was isolated only less frequently from infected groundnut root samples collected from Babile, Fedis and Gursum districts, whereas R. solani was isolated only from Babile and Fedis districts. Other fungi, such as Aspergillus, Penicillium, Trichoderma, and unidentified species were also recovered at very low rates, so they were presented as a group of “other fungi” (Table 3).

Pathogenicity test
Test for homogeneity of variance for the two trials was not significant (P = 0.8847); hence, the disease data for the two repeated trials were combined for analyses.
Pathogenicity tests indicated that all fungal species recovered from infected groundnut plants were able to causing root rots on groundnut var. Oldhale. Inoculated plants showed stunted growth and yellowing leaves. The root lesion on infected groundnut plants varied from light brown to black discoloration, and rotted root tissues were visually evident (Figure 3). In contrast, non-inoculated control groundnut plants were free from any visible disease symptoms. Significant variations (P < 0.0001) in disease severity were indicated among isolates of each fungal pathogen and among the five fungal pathogens tested (Figure 4). Variations in disease severity among three isolates of each fungal pathogen were significant, except for S. rolfsii. Mean disease severity index (SI) of the five fungal pathogens differed from 21.1 to 53.7%. Among these five fungal pathogens, F. solani (mean of 53.7%) was the most virulent in causing root rot on groundnut, followed by R. bataticola (46.7%) and F. oxysporum (33.7%), while R. solani (27.0%) and S. rolfisii (21.1%) were less virulent (Figure 4). All inoculated fungal pathogens were reisolated on artificial media, completing Koch’s postulates.

Interactive effect of pathogens
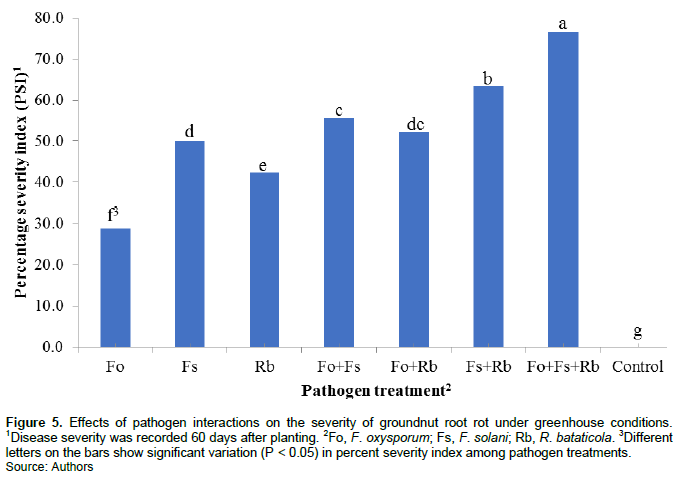
The effect of F. solani, F. oxysporum and R. bataticola singly and in combination on groundnut was studied in artificially-infested soil under greenhouse conditions. Test for homogeneity of variance for the two trials was not significant (P = 0.9723), thus disease data of the two repeat trials were combined for analyses. Results indicated that disease severity highly and significantly (P < 0.0001) varied with the pathogen treatments. Each pathogen alone caused significantly greater disease severity than the non-inoculated treatment. Root rot was more severe in soil infested with Fs+Rb (SI = 63.3%), Fs+Fo (55.6%), and Fo+Rb (52.2%) than either alone, but the combination of the three pathogens induced the greatest (76.7%) disease severity (Figure 5). This suggests that a synergistic relationship exists among these pathogens. During the reisolation of pathogens from infected groundnut roots, we often found that more than one pathogen was isolated at the same time on artificial media. This indicated that pathogens can share the same niche although the isolation rate was low for R. bataticola.
DISCUSSION
Groundnut root rot complex was found to be widespread in eastern Ethiopia although there was noticeable variation in disease intensity among the four districts (Babile, Darolebu, Fedis and Gursum) surveyed. The highest mean disease prevalence and incidence were recorded in Babile district, while the lowest was in Darolebu; incidence in Babile was greater by 8 to 29% than in other districts. This variation might be due to one or more of the following factors, including soil types, soil moisture content, inoculum density of the pathogens, other grower agricultural practices, and interaction between the host and pathogens (Tu, 1994; Palacios et al., 2014; Achuoth, 2018). Babile district relatively received the lowest rainfall and had high day temperatures during the survey seasons (Table 1), and these climatic conditions might be more conducive factors for disease development. Furthermore, most farmers in Babile had continuously grown groundnut year after year on the same field for several years due to its high market value as a cash crop and their land shortage.
This practice continues cultivation of groundnut on the same pieces of farm land may contribute to build-up of high fungal population in the soil, with the consequent increase in pathogen infection and disease development (Janila and Mula, 2015; Achuoth, 2018).
The survey data indicated that prevalence and incidence of groundnut root rot complex varied between cropping seasons, and greater disease prevalence and incidence were recorded during the 2015 cropping season than in 2014. In 2015, the seasonal rainfall was relatively low and erratic and had high day temperatures than in 2014 (Table 1). High temperature and low soil moisture might create an environment very favorable to the pathogens and their damage to groundnut. Root rot complex pathogens induce severe damage on groundnut under different environmental stresses like high temperature and drought-conditions (Wong et al., 1984; Rajamohan and Balabaskar, 2016), where disease incidence reached 95% in a few groundnut fields (Rojo et al., 2007). The effect of high temperature and low soil moisture on the intensity of groundnut root rots might be, at least partly, due to direct effect linked to increased pathogenic actions of the pathogens and/or partially due to moisture -and heat linked stresses forced on the plant predisposing to the pathogens. Symptoms caused by Fusarium spp. and R. bataticola are commonly observed during such conditions (Rajamohan and Balabaskar, 2016; Esmaeili-Taheri et al., 2017).
Among the pathogens in the groundnut root rot complex, F. oxysporum was found to be the most frequently isolated fungus in all four districts surveyed in eastern Ethiopia, followed by F. solani and R. bataticola (synonym = M. phaseolina). The current results consistent with the previous findings on the main groundnut root rot pathogens reported in Egypt (Atta-Alla et al., 2004), India (Rajamohan and Balabaskar, 2012), Pakistan (Zaman and Ahmed, 2012) and China (Li et al., 2014). The present results also in agreement with findings of Tarekegn et al. (2007), who previously isolated Fusarium spp. frequently from diseased groundnut roots in Babile district, part of which was covered in this survey. The findings, however, are in contrast with other previous findings in which R. solani appear to be the main groundnut root rot pathogen (Fath-ELBabe et al., 2017). Low isolation frequency of this pathogen in the present studies indicated its minor importance in eastern Ethiopia, and the prevailing dry conditions might be unfavorable for pathogen development. R. solani is favored by high moist condition, and is the predominant pathogen affecting groundnut in seasons of high rainfall (Porch et al., 2014).
The pathogenicity tests in the present study indicated that F. solani and R. bataticola were the most virulent and caused the most severe damage on groundnut among the fungi isolated from symptomatic groundnut roots, which are similar to findings reported by Elwakil and El-Metwally (2001), and Mahmoud (2015).
The F. oxysporum was the most common fungus recovered from diseased groundnut plants in current study. This fungus is omnipresent and considered to be associated mainly with wilt of several legume crops (Chopada et al., 2015; Ayele et al., 2021). However, reports from China indicated that F. oxysporum is a causal agent of groundnut root rots together with other fungal species (Li et al., 2014). In contrast, Sanogo and Puppala (2012) reported that F. oxysporum was either weakly pathogenic or a non-pathogenic species frequently isolated from groundnut roots; however, a previous study in Egypt (Atta-Alla et al., 2004) and the present study demonstrated that F. oxysporum is not only a prevalent fungal species, but also a pathogenic species causing severe root rot of groundnut. Groundnut plants inoculated with this fungus in the present study did not displayed disease symptoms that could be characterized as Fusarium wilts (Ayele et al., 2021). F. oxysporum enters its host crops through the roots, and causes either root rot or wilts when attacking plant vascular systems (Fravel et al., 2003; Farahani-Kofoet et al., 2020).
The pathogens interaction of F. solani, F. oxysporum and R. bataticola in causing a root rot complex of groundnut found to be synergistic, because disease severity was greater for groundnut plants in soil infested with the pathogen mixture than with either alone. Similar synergistic interactions have been reported for groundnut or other crops when infested by more than one pathogen (Frank, 1972; Willsey et al., 2018; Porto et al., 2019; Sumbul and Mahmood, 2020). Other studies demonstrate antagonism between R. solani and P. ultimum on bean (Pieczarka and Abawi, 1978), and F. oxysporum and R. solani on dry bean (de Jensen, 2000). Results of de Jensen (2000) showed that disease severity was reduced due to the occurrence of the two pathogens on the same plants; however, damage from the two pathogens enhanced when plant previously inoculated with one pathogen were consequently inoculated with other pathogen. The current study, however, demonstrated that simultaneous inoculation of F. solani, F. oxysporum, and R. bataticola resulted in the greatest root rot severity on groundnut; but not such antagonisms among the three tested fungal species were evident. This may partially be because all three fungal species in the current study were all together inoculated on groundnut plants; but more possible is a consequence of the fact that all three fungal species are present all together in soil (Porto et al., 2019), where fungal pathogens could resulted in enchased root rot severity when found together as a complex (Willsey et al., 2018; Fang et al., 2021).
CONCLUSION
Groundnut root rot complex was found to be widespread in East and West Haraghe zones, eastern Ethiopia, and prevalence and incidence of the disease varied among districts and between the two cropping seasons. The highest mean prevalence and incidence were recorded in the Babile district, followed by Gursum and Fedis. F. oxysporum was the most frequently isolated pathogen from diseased groundnut roots, followed by F. solani, and R. bataticola. During pathogenicity studies, these three pathogens also caused the most severe root rot on groundnut plants. Furthermore, the three pathogens combined treatment resulted in enhanced disease severity. The three pathogens are, therefore, the major causal agents of groundnut root rots in eastern Ethiopia. Further studies should be conducted in other groundnut growing areas of Ethiopia to get a clear picture of root rot etiology in Ethiopia. Furthermore, appropriate root rot management strategies, such as crop rotations, tillage practices, cultivar resistance, seed treatment, biological control methods and integrations, for the study area should be established.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
ACKNOWLEDGEMENTS
The authors appreciate the financial support of the Swedish Agency for Research Co-operation with Developing Countries (SIDA/SAREC), Madda Walabu University, and Association of African Universities. They thank farmers for allowing them to use their fields for disease assessment and sample collection. The authors also thank all staff members of Plant Pathology Laboratory of the School of Plant Sciences, Haramaya University, for their valuable support during the laboratory and greenhouse experiments.
REFERENCES
|
Achuoth MJ (2018). Evaluation of yields, pests and soil nutrients in sorghum/groundnut intercropping system in western Bahr, El Ghazal, Warrap and Abyei, South Sudan. International Journal of Agronomy and Agricultural Research 13(3):21-36. |
|
|
Ajeigbe HA, Waliyar F, Echekwu CA, Ayuba K, Motagi BN, Eniayeju D, Inuwa A (2014). A farmer's guide to groundnut production in Nigeria. Patancheru 502 324, Telangana, India: International Crops Research Institute for the Semi-Arid Tropics P 36. |
|
|
Amare K, Seltene A, Daniel E, Jemal A, Addisu G, Aliyi R, Yohanese P (2017). Registration of 'Babile-1', 'Babile-2', and 'Babile-3' groundnut varieties. East African Journal of Sciences 11(1):59-64. |
|
|
Atta-Alla SI, El-Samra IA, El-Korany AE, El-Sheikh MA, El-Nawam MF (2004). Management of the root rot of peanut in the newly reclaimed land in El-Behera Governorate, Egypt. Journal of Agriculture and Environmental Science of Alexandria University 3(1):9-26. |
|
|
Ayele TM, Gebremariam GD, Patharajan S (2021). Isolation, identification and In vitro test for the biocontrol potential of Trichoderma viride on Fusarium oxysporum f.sp. lycopersici. The Open Agriculture Journal 15(1):10-20. |
|
|
Bodah ET (2017). Root Rot Diseases in Plants: A review of common causal agents and management strategies. Agricultural Research and Technology: Open Access Journal 5(3):1-8. |
|
|
Burnett HL, Hunter BB (2003). Illustrated genera of imperfect fungi. 4th edition Burgess Pub. Co. Minneapolis, Minnesota, USA 218 p. |
|
|
Chala A, Abate B, Taye M, Mohammed A, Alemu T, Helge S (2014). Opportunities and constraints of groundnut production in selected drylands of Ethiopia. Drylands Coordination Group (DCG) Report No. 74, (March, 2014). Norway 50 p. |
|
|
Chopada GB, Singh P, Chandulal K (2015). Cultural and morphological variability among Fusarium oxysporum f.sp. lycopersici causing wilt of tomato in south Gujarat region. Archives of Phytopathology and Plant Protection 48(2):104-110. |
|
|
Central Statistical Agency (CSA) (2017). Report on area and production of major crops (Private peasant holdings, meher season). Ethiopian Agricultural Sample Survey, (2016/2017). Federal Democratic Republic of Ethiopia, CSA, Addis Ababa. |
|
|
de Jensen ECE (2000). Etiology and control of dry bean root rot in Minnesota. INIAP Archivo Historico Publishing, USA 396 p. |
|
|
District Agricultural Offices and National Meteorological Service Agency (2014, 2015). https://nimet.gov.ng/ |
|
|
Elwakil MA, El-Metwally MA (2001). Seed-borne fungi of peanut in Egypt: Pathogenicity and Transmission. Pakistan Journal of Biological Sciences 4:63-68. |
|
|
Esmaeili-Taheri A, Chatterton S, Foroud NA, Gossen BD, McLaren DL (2017). Identification and community dynamics of fungi associated with root, crown, and foot rot of field pea in western Canada. European Journal of Plant Pathology 147:489-500. |
|
|
Fang X, Zhang C, Wang Z, Duan T, Yu B, Jia X, Pang J, Ma L, Wang Y, Nan Z (2021). Co-infection by soil-borne fungal pathogens alters disease responses among diverse alfalfa varieties. Frontiers in Microbiology 12:664385. |
|
|
Food and Agriculture Organization/Statistics Database (FAOSTAT) (2022). Food and Agriculture Organization of the United Nations. FAO Statistical Databases of Crop Production (Accessed 18-03-2022). Available at: http://www.fao.org/faostat/en/#data/QCL |
|
|
Farahani-Kofoet RD, Witzel K, Graefe J, Grosch R, Zrenner R (2020). Species-specific impact of Fusarium infection on the root and shoot characteristics of Asparagus. Pathogens 9(6):1-20. |
|
|
Fath-ELBabe NMI, Elzaawely AA, Morsy SMA, El-Zahaby HM (2017). Control of peanut root rot using some chemical substances. Environment, Biodiversity and Soil Security 1:157-165. |
|
|
Frank ZR (1972). Pythium myriotylum and Fusarium solani as co-factors in a pod-rot complex of peanut. Phytopathology 62:1331-1334. |
|
|
Fravel D, Olivain C, Alabouvette C (2003). Fusarium oxysporum and its biocontrol. New Phytologist 157:493-502. |
|
|
Gezahagn K (2013). Economics of groundnut production in east Hararghe Zone of Oromia Regional State, Ethiopia. Science, Technology and Arts Research Journal 2 (2):135-139. |
|
|
Gomez KA, Gomez AA (1984). Statistical procedures for agricultural research, 2nd edition. John Willey and Sons, New York, USA. 680p. |
|
|
Hassuba MMM, El-Kholy RMA, El-Samadisy AM, Helalia AAR (2016). Evaluation of seed treatments with fungicides and bioagents in controlling of peanut diseases. Journal of Plant Protection and Pathology, Mansoura University 7(11):695-700. |
|
|
Janila P, Mula MG (2015). Cultural management practices of groundnut. International Crops Research Institute for the Semi-Arid Tropics 16 p. |
|
|
Janila P, Nigam SN, Pandey MK, Nagesh P, Varshney RK (2013). Groundnut improvement: use of genetic and genomic tools. Frontiers in Plant Science 4:23. |
|
|
Kassa Y, Beyene F, Haji J, Legesse B (2013). Impact of integrated soil and water conservation program on crop production and income in west Hararghe Zone, Ethiopia. International Journal of Environmental Monitoring and Analysis 1(4):111-120. |
|
|
Lamichhane JR, Venturi V (2015). Synergisms between microbial pathogens in plant disease complexes: a growing trend. Frontiers in Plant Science 6:385. |
|
|
Leslie JF, Summerell BA (2006). The Fusarium laboratory manual. Blackwell Publishing, Iowa, USA 387 p. |
|
|
Li X, Ding C, Zhang Y, Wang X (2014). Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biology and Biochemistry 72:11-18. |
|
|
Mahmoud EY, Eetmad-Draz EI, Abolela MF (2006). Evaluation of some peanut cultivars for the susceptibility of infection by damping-off, root and pod rot diseases and occurrence of aflatoxigenic fungi. Journal of Agricultural Science of Mansoura University 31(12):7589-7604. |
|
|
Mahmoud EY, Ibrahim MM, Essa TAA (2013). Efficacy of plant essential oils in controlling damping-off and root rots diseases of peanut as fungicides alternative. Journal of Applied Sciences Research 9(3):1612-1622. |
|
|
Mahmoud MA (2015). Efficiency of some bioagents and nemastop compound in controlling damping-off and root rot diseases on peanut plants. International Journal of Advanced Research in Biological Sciences 2(11):77-86. |
|
|
Meresa H, Tsehaye Y (2020). Interaction of phosphorus and foliar zinc on seed quality and Aspergillus infection on groundnut (Arachis hypogaea L.) genotypes in dryland area of Tanqua Abergelle, Ethiopia. International Journal of Life Sciences 8(1):59-69. |
|
|
Naseri B (2008). Root rot of common bean in Zanjan, Iran: major pathogens and yield loss estimates. Australasian Plant Pathology 37:546-551. |
|
|
Nelson PE, Toussoun TA, Marasas WFO (1983). Fusarium species: an illustrated manual for identification. Pennsylvania State University Press, University Park, 206p. |
|
|
Nordern AJ, Smith OD, Gorbet DW (1982). Breeding of cultivated peanut. In. Pattere HE, Youngs CT (eds.), Peanut Science and Technology. American Peanut Research and Education Society. Incorporated, Yoakun, Tex., pp. 95-122. |
|
|
Palacios S, Casasnovas F, Ramirez ML, Reynoso MM, Torres AM (2014). Impact of water potential on growth and germination of Fusarium solani soilborne pathogen of peanut. Brazilian Journal of Microbiology 45(3):105-1112. |
|
|
Pieczarka DJ, Abawi GS (1978). Effect of interactions between Fusarium, Pythium and Rhizoctonia on severity of bean root rot. Phytopathology 68:403-408. |
|
|
Porch TG, Valentin S, de Jensen CE, Beaver JS (2014). Identification of soil-borne pathogens in a common bean root rot nursery in Isabela, Puerto Rico. Journal of Agriculture of the University of Puerto Rico 98(1):1-14. |
|
|
Porto MAF, Ambrósio MMQ, Nascimento SRC, Cruz BLS, Torres RM (2019). Interaction of Fusarium solani, Macrophomina phaseolina and Rhizoctonia solani as root rot pathogens of Cucumis melo. Summa Phytopathologica 45(4):355-360. |
|
|
Rajamohan K, Balabaskar P (2016). In vivo evaluation of am fungi on dry root rot disease incidence and biometrics of groundnut. International Journal of Development Research 6(8):9074-9081. |
|
|
Rajamohan K, Balabaskar P (2012). Survey on the incidence of groundnut root rot disease in Cuddalore District of Tamil Nadu and assessing the cultural characters and pathogenicity of Macrophomina phaseolina (Tassi.) Goid. Asian Journal of Science and Technology 3(4):90-94. |
|
|
Rojo FG, Reynoso MM, Ferez M, Chulze SN, Torres AM (2007). Biological control by Trichoderma species of Fusarium solani causing peanut brown root rot under field conditions. Crop Protection 26:549-555. |
|
|
Sanogo S, Puppala N (2012). Microorganisms associated with valencia peanut affected by pod rot in New Mexico. Peanut Science 39:95-104. |
|
|
Sharma P, Saini MK, Deep S, Kumar V (2012). Biological control of groundnut root rot in farmer's field. Journal of Agricultural Science 4(8):48-59. |
|
|
Sumbul A, Mahmood I (2020). Interactive effect of Meloidogyne incognita and Macrophomina phaseolina on the development of root rot disease complex in relation to growth and physiological attributes of chickpea. Hellenic Plant Protection Journal 13:13-23. |
|
|
Tadesse E, Tucho TA, Hundesa F, Weldu G, Weldu T, Balay TK, Tibesso O (2014). Traditional cattle production in the highlands of Hararghe: Case study for east and west Zones of the highlands of Hararghe, eastern Ethiopia. Basic Research Journal of Agricultural Science and Review 3(12):122-130. |
|
|
Tarekegn G, Sakhuja PK, Swart WJ, Tamado T (2007). Integrated management of groundnut root rot using seed quality and fungicide seed treatment. International Journal of Pest Management 53(1):53-57. |
|
|
Tu J (1994). Effects of soil compaction, temperature, and moisture on the development of the Fusarium root rot complex of pea in southwestern Ontario. Phytoprotection 75(3):125-131. |
|
|
Wheeler JB (1969). An introduction to plant diseases. Wiley, London. 347 p. |
|
|
Willsey TL, Chatterton S, Heynen M, Erickson A (2018). Detection of interactions between the pea root rot pathogens Aphanomyces euteiches and Fusarium spp. using a multiplex qPCR assay. Plant Pathology 67:1912-1923. |
|
|
Wong DH, Barbetti MJ, Sivasithamparam K (1984). Effects of soil temperature and moisture on the pathogenicity of fungi associated with root rot of subterranean clover. Australian Journal of Agricultural Research 35:675-684. |
|
|
Zaman N, Ahmed S (2012). Survey of root rot of groundnut in rainfed areas of Punjab, Pakistan. African Journal of Biotechnology 11(21):4791-4794. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0