Full Length Research Paper
ABSTRACT
Soil microbial communities play a vital role in ecosystem functioning by enhancing mineral nutrition and protecting forest trees against pathogens through mycorrhizal symbiosis. However, knowledge of the diversity and assemblage of belowground fungal communities associated with native host trees in tropical Africa is incomplete. Using high-throughput sequencing, this study examined soil fungal communities in the rhizosphere of five ectomycorrhizal trees (EcM) from (5) countries using ITS and LSU regions. Unconstrained ordination of fungal species was performed using principal component analysis based on their EcM tree rhizosphere affiliation. The ANOSIM test assessed the similarity between the fungal community composition associated with the EcM trees. Overall, 90 species belonging to 84 genera, 71 families, 40 orders and 4 phyla were identified. Soil fungal communities were host specific (P = 0.001). Basidiomycota were more frequently observed in the rhizosphere of Fabaceae, except for I. doka, whereas Ascomycota are more abundant in the rhizosphere of Phyllanthaceae (U. togoensis) and Dipterocarpaceae (M. kerstingii). The genus Sebacina is predominantly linked to M. kerstingii and I. tomentosa, while Russula is dominant under B. grandiflora and, Inocybe with I. tomentosa. This study provides new insights into in the rhizosphere of native forest trees in West Africa and highlights areas for future research.
Key words: DNA metabarcoding, ectomycorrhizal association, molecular species, Soil microorganisms, soil fungi, timber trees.
INTRODUCTION
The rhizosphere is considered to be the narrow zone of soil immediately surrounding plant roots (Marschner et al., 2004; Olahan et al., 2016). This area is home to a wide range of interactions between plant roots and microorganisms, which affect soil physical, chemical, and biological processes that sustain biodiversity and ecosystems (Nihorimbere et al., 2011; Sathya et al., 2016; Lu et al., 2018). A major group of microorganisms found in the rhizosphere are fungi, responsible in part for colonizing the roots of a plethora of plant species (Olahan et al., 2016; Sathya et al., 2016; Dlamini et al., 2022). Rhizospheric fungi play a vital role in the soil food chain, participating in the recycling of soil carbon and nutrients (Larekeng et al., 2019; Pattnaik and Busi, 2019), and the transformation of hard-to-digest organic matter (such as lignin and other soil organic matter) into usable forms for other organisms (Stokland et al., 2012; Grzyb et al., 2021). Through enzymatic activities, fungal hyphae physically bind soil particles together, creating stable aggregates that contribute to increased soil aeration, water infiltration, and water holding capacity of the soil, thereby enhancing soil resistance to erosion (Vogelsang et al., 2004; van der Wal et al., 2009). As a result, rhizospheric fungi are directly involved in soil fertility (Sterkenburg et al., 2015; Rashid et al., 2016) and contribute to the mitigation of soil degradation (Rashid et al., 2016; Rosas-Medina et al., 2020).
Among rhizospheric fungi, mycorrhizal fungi comprise one of the major groups since they are associated with more than 90% of known terrestrial plants (Smith and Read, 2008; Nilsson et al., 2019; Islam et al., 2022). Mycorrhizal fungi significantly improve the absorption and use of nutrients by host plants, stimulate growth, increase stress and disease resistance, and thereby contribute to maintaining the aboveground primary productivity of forest and ecosystem stability (Larekeng et al., 2019; Thind et al., 2022). According to root morphological differentiation, there are many types of mycorrhizal fungi of which one of them is ectomycorrhizal (EcM) fungi. They are obligate partners of most woody plant species that majorly belong to the families Fagaceae, Dipterocarpaceae, Phyllanthaceae, Myrtaceae, etc. (Brundrett and Tedersoo, 2018; Corrales et al., 2018). In tropical Africa, some EcM trees that belong to these families are Afzelia africana Smith ex Persoon, Berlinia grandifolia (Vahl) Hutch. and Dalziel, Monotes kerstingii Gilg, Isoberlinia doka Craib and Stapf, Isoberlinia tomentosa (Harms) Craib and Stapf, Uapaca togoensis Pax, etc. (Bâ et al., 2012; Houdanon et al., 2019). They are economically important trees and because of their socio-economic value, these species are facing major pressure from the local population, including charcoal production, and illegal logging for furniture (Balima et al., 2018; Mohammed et al., 2021). In addition, natural regeneration is not able to compensate for the removal of trees from the forest. Therefore, attempts to plant nursery-produced seedlings in the wild have been considered (Ogbimi et al., 2020; Ogbimi and Sakpere, 2021). However, since nursery production does not include knowledge of the niche of these plant species in their natural habitats, the results of planting in the wild are not satisfactory. Given that fungi play a key role in plant growth and health, there is a clear need to better understand the soil mycobiome surrounding native forest trees to develop an effective sustainable management strategy.
Until recently, studies on fungal diversity in West Africa have relied primarily on fruiting bodies surveys, mycelia isolations, and spore identification (Straatsma et al., 2001; Luo et al., 2020). Fruit bodies-based surveys do not allow a total evaluation of the fungal community (Kubartová et al., 2012; Shirouzu et al., 2016), because even if a fungus has basidiomata large enough to be spotted, they may go unnoticed because fruiting body formation is both seasonal and ephemeral (Shirouzu et al., 2016). Many taxa such as mycorrhizal and parasitic fungi may not grow or produce reproductive structures on artificial media even if they are potentially culturable (Allen et al., 2003; Senanayake et al., 2020). In addition to the aforementioned methods, spore identification is traditionally used to identify the rhizosphere arbuscular mycorrhizal fungi (Rodríguez-Morelos et al., 2014; Xavier and Rodrigues, 2020). However, even though this method is important in fungal taxonomy, it is time- and energy-consuming and susceptible to variability in spore morphology description, because host species and microbial age may be very challenging to differentiate spores of similar species (Bhat et al., 2014; Senanayake et al., 2020). Recent studies using high-throughput sequencing of environmental samples have greatly improved our understanding of the community and diversity of rhizosphere soil fungi (Tedersoo et al., 2014; Qin, 2018; Zhu et al., 2018; Nilsson et al., 2019; Tremblay et al., 2019; Meidl et al., 2021).
One of the most accepted methods for high throughput sequencing is the generation of the amplicon sequence variants (ASVs). So far, this method has been mainly used to study soil mycobiome in temperate and boreal regions (Wu et al., 2019; Lance et al., 2020; Rosas-Medina et al., 2020), while very few studies have comprehensively assessed the diversity, and community composition of soil fungi in tropical African forest ecosystems (Meidl et al., 2021). Here, PacBio sequencing was employed to assess the diversity and community composition of fungi found in the rhizosphere of five West African native trees.
MATERIALS AND METHODS
Study area
The soil samples used in this study were collected across five West African countries namely Benin, Burkina Faso, Guinea, Côte d’Ivoire, and Mali. In total, nine forests containing at least one of the targeted EcM tree species were selected. The different forests range from woodlands to gallery forests: The gallery forests and the woodland of Kota in Benin, the Kou gallery forest and the Niangoloko forest reserve in Burkina-Faso, the Farako1 forest reserve and the Farako15 forest reserve in Mali, the Bissandougou forest reserve and Moussaya forest reserves in Guinae and the Kouadianikro gallery forest in Côte d’Ivoire (Figure 1).
Sampling design and methods
Within each study site, we established a plot of 50 m × 50 m (2500 m2) in woodlands and a rectangular plot of 30 m × 80 m (2,400 m2) within gallery forests due to their shape. Within each plot, five EcM trees were targeted, namely I. doka, I. tomentosa, U. togoensis, M. kerstingii, and B. grandiflora. Ten trees were chosen in proportion to their abundance, while ensuring that each of the EcM trees in the plot is represented at least once and that all sampled trees were at least eight meters apart. Under each targeted tree, two soil samples of about 200 g around 1 m was taken from each side of the trunk using a small shovel to collect the first 15 cm of soil. The two soil samples were pooled in a plastic bag. A total of 90 (5 EcM trees x 2 samples x 9 sites) soil samples were collected at a rate of 10 samples per site. Later on, the collected soil samples were processed following the protocol described by Tedersoo et al. (2014).
DNA extraction, sequencing and bioinformatics analyses
For the DNA extraction and sequencing, soil samples were sent to the Department of Ecology and Genetics, Evolutionary Biology, Uppsala University. A subsample of approximately 250 mg was placed in a separate 2.0 ml tube containing 750 µl of field lysis and preservation buffer (Xpedition Soil/Fecal DNA miniprep, Zymo Research Corporation, Irvine, California, USA) and lysed in the field using a portable bead beater (TeraLyser, Zymo Research Corporation).
Extraction, amplification, sequencing, and clustering of sequences into amplicon sequence variants (ASVs) were performed as described by Meidl et al. (2021). For more details, see the methodology of Meidl et al. (2021). The taxonomic attribution of the different ASVs was carried out on the PlutoF platform (Kõljalg et al., 2019) using the PROTAX software (Somervuo et al., 2016) (publication date 2020-10-21), configured by the Index Fungorum taxonomic database and the UNITE reference sequence database (Nilsson et al., 2016). We recorded for each query ASV the most likely taxonomic identity at the phylum, class, order, family, genus, and species levels, as well as the uncertainty of these assignments, measured by probabilistic placement. The authors note that the PROTAX uncertainty estimates explain the possibility that the species is unknown to science (that is, not included in the taxonomic database), or known to science but lacking sequences reference (Somervuo et al., 2016; Abarenkov et al., 2018).
Data processing and analysis
To illustrate the fungal taxonomic composition associated with the rhizosphere of the target EcM trees, we constructed a Krona wheel for each tree using Protax-fungi in PlutoF platform from ASV diversity. Alpha diversity was determined for each EcM tree by calculating species richness and the Shannon diversity index. The similarity analysis (ANOSIM) was used to assess the similarity between the fungal communities associated with EcM trees. Through principal component analysis, we highlight fungal species affiliation with each EcM tree, and to identify the potential fungal species which better characterize each EcM tree. Finally, the Jaccard similarity index was calculated to compare the proportion of species shared by different EcM trees. All these analyses were carried out using the vegan package (Oksanen et al. 2022) with the statistical software R version 3.6.2 (R Core Team, 2019) and the ggplot2 package (Wickham, 2016) was used to create the nMDS graph.
RESULTS
Taxonomic composition of fungal communities associated with the rhizosphere of targeted EcM trees
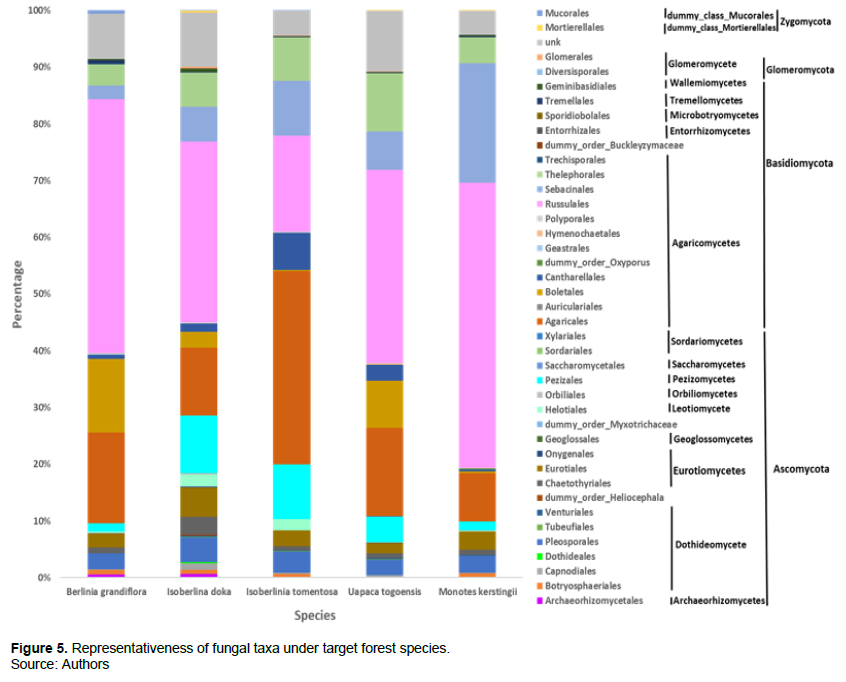
Grouping the sequences into amplicon sequence variants (ASVs) gave a total of 1147 ASVs. In sum, 1051 ASV (91.63%) were identified as fungi. On the Krona wheels (Figures 2 to 4, Supplementary materials A, B, C, D and E for more detail), the color scales show the type and confidence level of each taxonomic placement. Color scales 1 to 3 correspond to the identified taxonomic units for which the proportion of reliable identifications ranges from 50… 100% (1), 0… 50% (2) or 0 % Color 3. Scales 4 to 6 correspond to unknown taxonomic units. In total, four taxonomic groups of fungi such as Basidiomycota, Ascomycota, Glomeromycota, Zygomycota were identified from the rhizosphere of the targeted EcM trees.
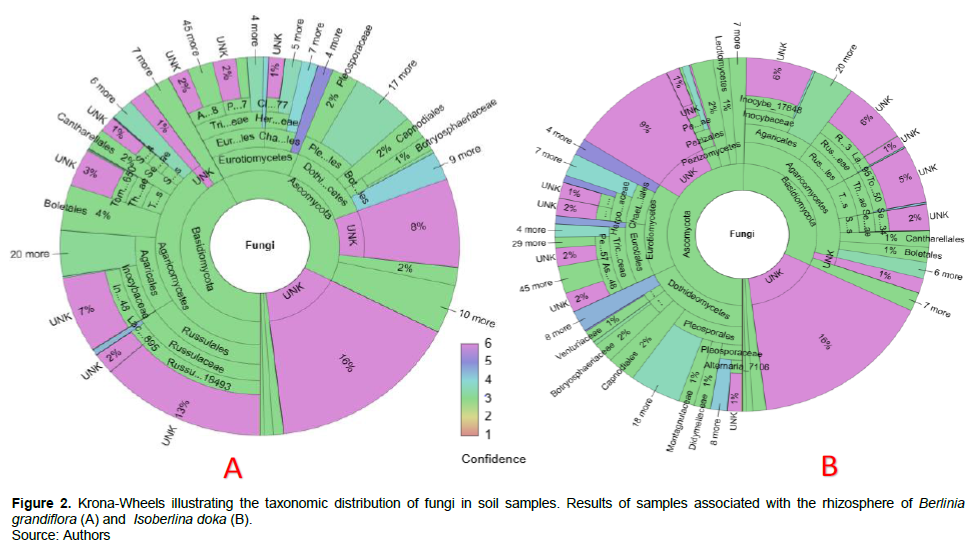
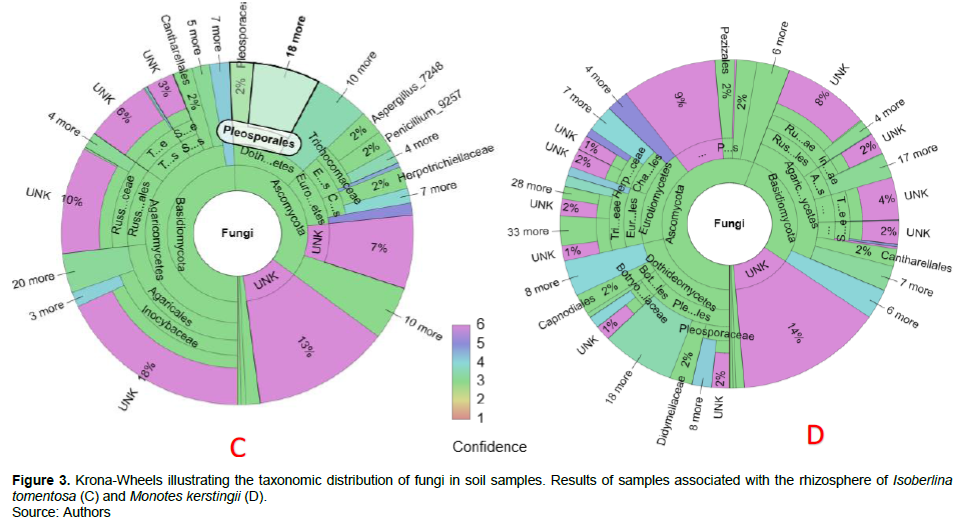
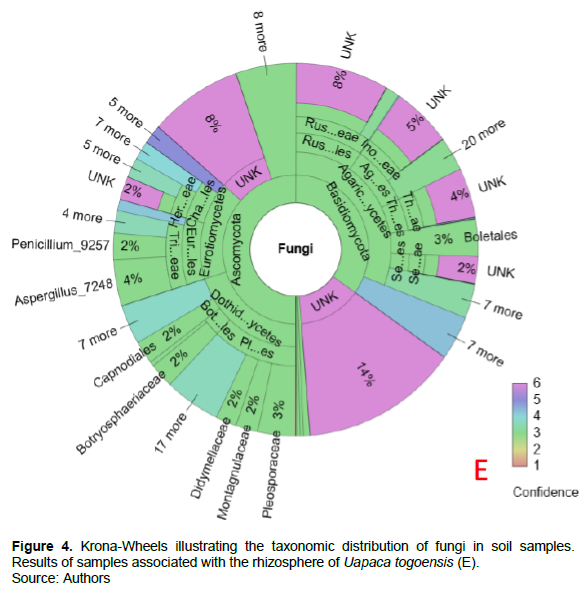
These latter are unevenly distributed for each EcM tree. For example, Basidiomycota are most dominant under B. grandiflora (42%) and I. tomentosa (49%); while Ascomycota are the most dominant under I. doka (50%), M. kerstingii (56%), and U. togoensis (50%). Glomeromycota and Zygomycota are weakly represented under the target EcM trees. Sixteen percent of the sequences associated with B. grandiflora and I. doka are unidentified or unknown. Fourteen percent of the fungi sampled under M. kerstingii and U. togoensis are unidentified, whilst unknown taxa make up to 13% of total fungal community under I. tomentosa.
In general, Russulales is the most dominant fungal group under B. grandiflora, I. doka, U. togoensis, and M. kerstingii, while Agaricales is more abundant in the rhizosphere of I. tomentosa (Figure 5). Sebacinales are more represented under M. kerstingii than the other trees investigated in contrast to Boletales, which are more represented under B. grandifolia. I. doka and I. tomentosa have the highest proportion of Pezizales. Cantharellales, an important group of edible fungi in tropical Africa, is best represented under I. tomentosa.
Genera representativeness under the different forest species
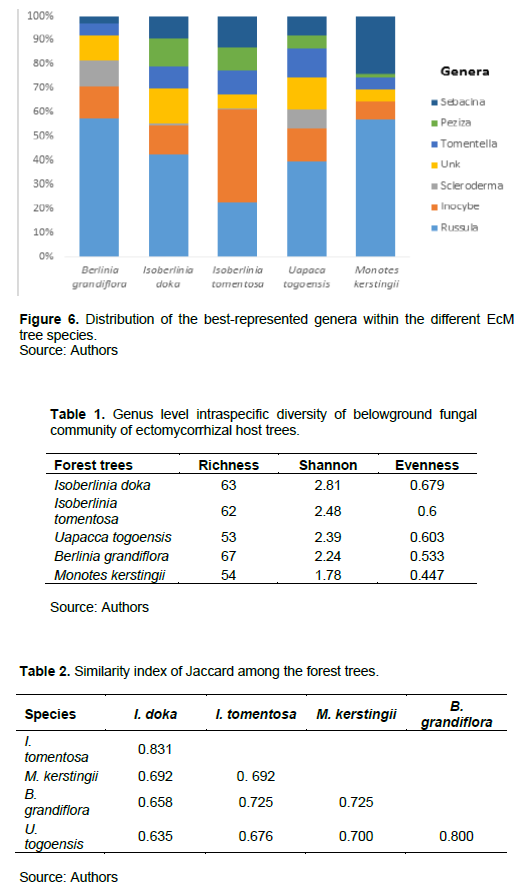
A total of 1051 ASV, including 810 (77.07%) belonging to 90 species from 84 genera, 71 families, 40 orders, 19 classes, and 04 phyla have been recorded. Moreover, 66.67% of this specific richness is observed under B. grandiflora (60 species), against 62.22% for I. doka (56 species), 53.33% for I. tomentosa (48 species), 48.89% for U. togoensis (44 species), and 47.78% for M. kerstingii (43 species). The real diversity is probably much higher because about 60% of the genera (50 genera for all EcM trees combined) could not be identified up to species level. About 22.93% (241) of the ASV remained unidentified and were not included in this analysis. Russula is better represented under B. grandiflora, I. doka, U. togoensis, and M. kerstingii unlike Inocybe that is much more observed under I. tomentosa (Figure 6).
Diversity of belowground fungal communities of five EcM trees
Table 1 presents the intraspecific diversity of the belowground fungal communities of the different tree species in the EcM. At the genus level, the belowground fungal communities were found to be the most diverse for Isoberlinia doka (G = 63, H' = 2.81, J = 0.679) and the least diverse for Monotes kerstingii (G = 54, H' = 1.78, J = 0.447). On the other hand, fungal generic diversity affiliated with Uapaca togoensis (G = 53, H' = 2.39, J = 0.603) was approximately equal to that of Isoberlinia tomentosa (G = 62, H' = 2.48, J = 0.6).
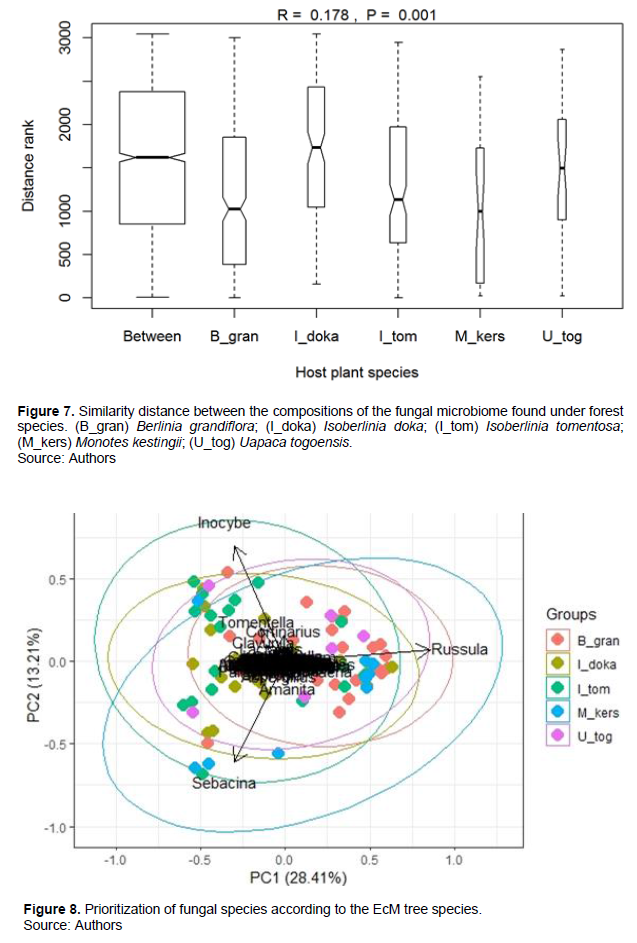
Considering pairwise EcM trees, Jaccard's similarity index (Table 2) indicated generally large proportions of shared fungal genera. Indeed, similarity (0.635) was obtained between I. doka and U. togoensis; but I. doka and I. tomentosa shared the largest number of taxa (Jaccard index = 0.831). Although the proportion of genera shared was greater than 0.6 in all pairwise cases, the similarity analysis (ANOSIM) supported the evidence that at the genus level, the belowground fungal community associated with the rhizosphere of at least one of the five EcM trees differed significantly from the others (P < 0.05, Figure 7).
Categorization of below-ground fungal species according to EcM hosts
Figure 8 presents the projection of fungal genera generated for each EcM tree according to the principal components 1 and 2. Figure 8 suggests that EcM trees hardly cluster separately and share a large number of fungal genera as the similarity index of Jaccard indicated it. This makes it difficult to clearly identify the genera that characterized the fungal community of each tree. Nevertheless, through the projection of the circles, the genus Russula seems to cluster more with B. grandiflora; while Sebacina seems more associated with M. kerstingii and I. tomentosa; and the genus Inocybe clusters more with I. tomentosa.
DISCUSSION
CONCLUSION
Until recently, estimates of total fungal diversity did not include results from large-scale environmental sequencing methods, especially in West African regions. This study constitutes the first major exploration of the edaphic fungal communities of West African ecosystems, revealing insufficient sampling effort in currently neglected ecosystems and regions. The authors’ data provide a baseline for phylogenetic placement and taxonomic resolution of environmental sequences of five EcM trees of socio-economic importance in West Africa.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
ACKNOWLEDGEMENTS
The authors would like to thank the Department of Ecology and Genetics, Evolutionary Biology, Uppsala University for molecular analyzes and the German Federal Ministry of Education and Research (BMBF- Germany) grant agreement 01DG20015.
FUNDING
Sampling was funded by the National Geographic Society as part of exploration grant #CP-126R-17.
REFERENCES
|
Abarenkov K, Somervuo P, Nilsson RH, Kirk PM, Huotari T, Abrego N, Ovaskainen O (2018). Protax-fungi: a web-based tool for probabilistic taxonomic placement of fungal internal transcribed spacer sequences. New Phytologist 220:517-525. |
|
|
Allen MF, Swenson W, Querejeta JI, Egerton-Warburton LM, Treseder KK (2003). Ecology of Mycorrhizae:A Conceptual Framework for Complex Interactions among Plants and Fungi. Annual Review of Phytopathology 41:271-303. |
|
|
Bâ AM, Duponnois R, Moyersoen B, Diédhiou AG (2012). Ectomycorrhizal symbiosis of tropical African trees. Mycorrhiza 22:1-29. |
|
|
Balima LH, Nacoulma BMI, Ekué MRM, Kouamé FNG, Thiombiano A (2018). Use patterns, use values and management of Afzelia africana Sm. in Burkina Faso:Implications for species domestication and sustainable conservation. Journal of Ethnobiology and Ethnomedicine 14:1-14. |
|
|
Bhat BA, Sheikh MA, Tiwari A (2014). Impact of various edaphic factors on AMF spore population and diversity in Catharanthus roseus at Gwalior. International Journal of Plant Sciences 9:1-6. |
|
|
Brundrett MC, Tedersoo L (2018). Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytologist 220:1108-1115. |
|
|
Burke DJ, López-Gutiérrez JC, Smemo KA, Chan CR (2009). Vegetation and soil environment influence the spatial distribution of root-associated fungi in a mature beech-maple forest. Applied and Environmental Microbiology 75:7639-7648. |
|
|
Corrales A, Henkel TW, Smith ME (2018). Ectomycorrhizal associations in the tropics - biogeography, diversity patterns and ecosystem roles. New Phytologist 220:1076-1091. |
|
|
Crous PW, Rong IH, Wood A, Lee S, Glen H, Botha W, Slippers B, De Beer WZ, Wingfield MJ, Hawksworth DL (2006). How many species of fungi are there at the tip of Africa? Studies in Mycology 55:13-33. |
|
|
Diédhiou AG, Selosse MA, Galiana A, Diabaté M, Dreyfus B, Bâ AM, de Faria SM, Béna G (2010). Multi-host ectomycorrhizal fungi are predominant in a Guinean tropical rainforest and shared between canopy trees and seedlings. Environmental Microbiology 12:2219-2232. |
|
|
Djotan AKG, Matsushita N, Vaario LM, Yorou NS, Fukuda K (2021). Arbuscular mycorrhizas in the roots of afzelia africana, Entada Africana, and Pterocarpus Erinaceus. Applied Ecology and Environmental Research 19:833-848. |
|
|
Dlamini SP, Akanmu AO, Babalola OO (2022). Rhizospheric microorganisms: The gateway to a sustainable plant health. Frontiers in Sustainable Food Systems 6. |
|
|
Ebenye MHC, Taudière A, Niang N, Ndiaye C, Sauve M, Awana NO, Verbeken M, De Kesel A, Séne S, Diédhiou AG, Sarda V, Sadio O, Cissoko M, Ndoye I, Selosse MA, Bâ AM (2017). Ectomycorrhizal fungi are shared between seedlings and adults in a monodominant Gilbertiodendron dewevrei rain forest in Cameroon. Biotropica 49:256-267. |
|
|
Egger KN (1986). Substrate hydrolysis patterns of post-fire Ascomycetes (Pezizales). Mycological Society of America 78:771-780. |
|
|
Fonton NH, Atindogbe G, Fandohan B, Lejeune P, Ligot G (2012). Structure spatiale des arbres des savanes boisées et forêts claires soudaniennes:Implication pour les enrichissements forestiers. Biotechnology, Agronomy and Society and Environment 16:429-440. |
|
|
Gebhardt S, Neubert K, Wöllecke J, Münzenberger B, Hüttl RF (2007). Ectomycorrhiza communities of red oak (Quercus rubra L.) of different age in the Lusatian lignite mining district, East Germany. Mycorrhiza 17:279-290. |
|
|
Giollant M, Guillot J, Damez M, Dusser M, Didier P, Didier É (1993). Characterization of a lectin from Lactarius deterrimus. Plant Physiology 101:513-522. |
|
|
Gorzelak MA, Asay AK, Pickles BJ, Simard SW (2015). Inter-plant communication through mycorrhizal networks mediates complex adaptive behaviour in plant communities. AoB Plants 7:plv050. |
|
|
Grzyb A, Wolna-Maruwka A, Niewiadomska A (2021). The significance of microbial transformation of nitrogen compounds in the light of integrated crop management. Agronomy 11 p. |
|
|
Henry C, Selosse M-A, Richard F, Ramanankierana H, Ducousso M (2021). Comprendre la dynamique des communautés mycorhiziennes lors des successions végétales. Deuxième partie?:Potentialités d ' applications à la restauration des écosystèmes forestiers (revue bibliographique ) To cite this version?:HAL Id?:hal-03447480 Co. Hal open science:125-150. |
|
|
Houdanon R, Tchan I, Laourou G, Codjia J, Badou S, Aignon L, Boni S, Yorou N (2019). Spatial Structure Of Ectomycorrhizal Trees In Wooded Savannas Of Guineo-Sudanian Ecozone In West Africa. Tropical forest science 31:1-11. |
|
|
Houngnandan P, Yemadje R, Kane A, Boeckx P, Van Cleemput O (2009). Les glomales indigènes de la forêt claire à Isoberlinia doka (Craib et Stapf) à Wari-Maro au centre du Bénin. Tropicultura 27:83-87. |
|
|
Ishida TA, Nara K, Hogetsu T (2007). Host effects on ectomycorrhizal fungal communities:Insight from eight host species in mixed conifer-broadleaf forests. New Phytologist 174:430-440. |
|
|
Islam M, Al-Hashimi A, Ayshasiddeka M, Ali H, El Enshasy HA, Dailin DJ, Sayyed RZ, Yeasmin T (2022). Prevalence of mycorrhizae in host plants and rhizosphere soil:A biodiversity aspect. PLoS ONE 17:1-14. |
|
|
Johnson D, Vandenkoornhuyse P, Leake JR, Gilbert L, Booth RE, Grime JP, Young JPW, Read DJ, Booth E, Leakel R, Gilbert L, Johnsonl D, Vandenkoornhuyse PJ, Young W, Read DJ, Peter J, Grime JP (2004). Plant communities affect arbuscular mycorrhizal fungal diversity and community composition in grassland microcosm. New Phytologist 161:503-515. |
|
|
Kõljalg U, Abarenkov K, Zirk A, Runnel V, Piirmann T, Pöhönen R, Ivanov F (2019). PlutoF:Biodiversity data management platform for the complete data lifecycle. Biodiversity Information Science and Standards 3:1-5. |
|
|
Kretzer A, Li Y, Szaro T, Bruns TD (1996). Internal transcribed spacer sequences from 38 recognized species of Suillus sensu lato: Phylogenetic and taxonomic implications. Mycologia 88:776-785. |
|
|
Kubartová A, Ottosson E, Dahlberg A, Stenlid J (2012). Patterns of fungal communities among and within decaying logs, revealed by 454 sequencing. Molecular Ecology 21(18):4514-4532. |
|
|
Lance AC, Burke DJ, Hausman CE, Burns JH (2020). High-throughput sequencing provides insight into manipulated soil fungal community structure and diversity during temperate forest restoration. Restoration Ecology 28:S365-S372. |
|
|
Larekeng SH, Gusmiaty, Restu M, Tunggal A, Susilowati A (2019). Isolation and identification of rhizospheric fungus under Mahoni (Swietenia mahagoni) stands and its ability to produce IAA (Indole Acetid Acid) hormones. IOP Conference Series: Earth and Environmental Science 343. |
|
|
LeDuc SD, Lilleskov EA, Horton TR, Rothstein DE (2013). Ectomycorrhizal fungal succession coincides with shifts in organic nitrogen availability and canopy closure in post-wildfire jack pine forests. Oecologia 172:257-269. |
|
|
Lu T, Ke M, Lavoie M, Jin Y, Fan X, Zhang Z, Fu Z, Sun L, Gillings M, Peñuelas J, Qian H, Zhu YG (2018). Rhizosphere microorganisms can influence the timing of plant flowering. Microbiome 6:1-12. |
|
|
Luo Y, Wang Z, He Y, Li G, Lv X, Zhuang L (2020). High-throughput sequencing analysis of the rhizosphere arbuscular mycorrhizal fungi (AMF) community composition associated with Ferula sinkiangensis. BMC Microbiology 20:1-14. |
|
|
Marschner P, Crowley D, Yang CH (2004). Development of specific rhizosphere bacterial communities in relation to plant species, nutrition and soil type. Plant and Soil 261:199-208. |
|
|
Massicotte HB, Molina R, Luoma DL, Smith JE (1994). Biology of the ectomycorrhizal genus, Rhizopogon II. Patterns of host?fungus specificity following spore inoculation of diverse hosts grown in monoculture and dual culture. New Phytologist 126:677-690. |
|
|
McGuire KL (2007). Common ectomycorrhizal networks may maintain monodominance in a tropical rain forest. Ecology 88(3):567-574. |
|
|
Meidl P, Furneaux B, Tchan KI, Kluting K, Ryberg M, Guissou ML, Soro B, Traoré A, Konomou G, Yorou NS, Rosling A (2021). Soil fungal communities of ectomycorrhizal dominated woodlands across West Africa. MycoKeys 81:45-68. |
|
|
Mohammed EMI, Elhag AMH, Ndakidemi PA, Treydte AC (2021). Anthropogenic pressure on tree species diversity, composition, and growth of balanites aegyptiaca in dinder biosphere reserve, Sudan. Plants 10:1-18. |
|
|
Molina R, Trappe J (1994). Biology of the ectomycorrhizal genus, Rhizopogon I. Host associations, host?specificity and pure culture syntheses. New Phytologist 126:653-675. |
|
|
Nihorimbere V, Ongena M, Smargiassi M, Thonart P (2011). Effet bénéfique de la communauté microbienne de la rhizosphère sur la croissance et la santé des plantes. Biotechnology, Agronomy and Society and Environment 15:327-337. |
|
|
Nilsson RH, Wurzbacher C, Bahram M, Coimbra VRM, Larsson E, Tedersoo L, Eriksson J, Ritter CD, Svantesson S, Sánchez-García M, Ryberg M, Kristiansson E, Abarenkov K (2016). Top 50 most wanted fungi. MycoKeys 12:29-40. |
|
|
Nilsson RH, Anslan S, Bahram M, Wurzbacher C, Baldrian P, Tedersoo L (2019). Mycobiome diversity:high-throughput sequencing and identification of fungi. Nature Reviews Microbiology 17:95-109. |
|
|
Ogbimi ER, Sakpere AMA (2021). Germination and seedling growth in Afzelia africana Sm. ex. Pers. Ife Journal of Science 23:41-50. |
|
|
Ogbimi ER, Sakpere AM, Akinropo SM (2020). Vegetative propagation of Afzelia africana Sm. Ex Pers.:a multipurpose and threatened tree. International Journal of Biological and Chemical Sciences 14:204-212. |
|
|
Oksanen J, Simpson GL, Blanchet FG, Solymos P, Stevens MHH, Szoecs E, Wagner H, Barbour M, Bedward M, Bolker B, Borcard D, Carvalho G, Chirico M, Durand S, Beatriz H, Evangelista A, Friendly M, Hannigan G, Hill MO, Lahti L, Mcglinn D, Ribeiro E, Smith T, Stier A, Ter CJF (2022). Community Ecology Package. version 2.9 (2013):1-295. |
|
|
Olahan G, Sule I, Garuba T, Salawu Y (2016). Rhizosphere and Non-Rhizosphere Soil Mycoflora of Corchorus Olitorius (Jute). Science World Journal 11(3):23-26. |
|
|
Pattnaik SS, Busi S (2019). Rhizospheric Fungi:Diversity and Potential Biotechnological Applications. In:Yadav A, Mishra S, Singh S, Gupta A (eds) Recent Advancement in White Biotechnology Through Fungi. Fungal Biology 73-100. |
|
|
Qin Y (2018). 2 University of Oklahoma Studying microbial community diversities using high-throughput techniques and computational tools. Available from: https://shareok.org/bitstream/handle/11244/316306/2018_Qin_Yujia_Dissertation.pdf?sequence=2&isAllowed=y |
|
|
Rashid MI, Mujawar LH, Shahzad T, Almeelbi T, Ismail IMI, Oves M (2016). Bacteria and fungi can contribute to nutrients bioavailability and aggregate formation in degraded soils. Microbiological Research 183:26-41. |
|
|
Rodríguez-Morelos VH, Soto-Estrada A, Pérez-Moreno J, Franco-Ramírez A, Díaz-Rivera P (2014). Arbuscular mycorrhizal fungi associated with the rhizosphere of seedlings and mature trees of Swietenia macrophylla (Magnoliophyta : Meliaceae) in Los Tuxtlas, Veracruz, Mexico. Revista Chilena de Historia Natural 87:1-10. |
|
|
Rosas-Medina M, Maciá-Vicente JG, Piepenbring M (2020). Diversity of Fungi in Soils with Different Degrees of Degradation in Germany and Panama. Mycobiology 48:20-28. |
|
|
Sathya A, Vijayabharathi R, Gopalakrishnan S (2016). Microbial inoculants in sustainable agricultural productivity: Vol. 2: Functional applications. Microbial Inoculants in Sustainable Agricultural Productivity: Volume 2: Functional Applications:1-308. |
|
|
Senanayake I, Rathnayaka A, Marasinghe D, Calabon M, Gentekaki E, Lee H, Hurdeal V, Pem D, Dissanayake L, Wijesinghe S, Bundhun D, Goonasekara I, Abeywickrama P, Bhunjun C, Jayawardena R, Wanasinghe DN, Jeewon R, Bhat DJ, Xiang M (2020). Morphological approaches in studying fungi:collection, examination, isolation, sporulation and preservation. Mycosphere 11:2678-2754. |
|
|
Shirouzu T, Uno K, Hosaka K, Hosoya T (2016). Early-diverging wood-decaying fungi detected using three complementary sampling methods. Molecular Phylogenetics and Evolution 98:11-20. |
|
|
Smith SE, Read DJ (2008). Mycorrhizal Symbiosis. Third. Academic Press, Cambridge. |
|
|
Somervuo P, Koskela S, Pennanen J, Henrik Nilsson R, Ovaskainen O (2016). Unbiased probabilistic taxonomic classification for DNA barcoding. Bioinformatics 32:2920-2927. |
|
|
Sterkenburg E, Bahr A, Brandström Durling M, Clemmensen KE, Lindahl BD (2015). Changes in fungal communities along a boreal forest soil fertility gradient. New Phytologist 207:1145-1158. |
|
|
Stokland JN, Siitonen J, Jonsson BG (2012). Biodiversity in Dead Wood. Cambridge University Press, Cambridge. |
|
|
Straatsma G, Ayer F, Egli S (2001). Species richness, abundance, and phenology of fungal fruit bodies over 21 years in a Swiss forest plot. Mycological Research 105:515-523. |
|
|
Taylor DL, Bruns TD (1997). Independent, specialized invasions of ectomycorrhizal mutualism by two nonphotosynthetic orchids. Proceedings of the National Academy of Sciences USA 94:4510-4515. |
|
|
Taylor DL, Bruns TD, Leake JR, Read DJ (2002). Mycorrhizal Specificity and Function in Myco-heterotrophic Plants. Mycorrhizal Ecology 157:375-413. |
|
|
Tedersoo L, Smith ME (2013). Lineages of ectomycorrhizal fungi revisited: Foraging strategies and novel lineages revealed by sequences from belowground. Fungal Biology Reviews 27:83-99. |
|
|
Tedersoo L, Smith ME (2017). Ectomycorrhizal Fungal Lineages:Detection of Four New Groups and Notes on Consistent Recognition of Ectomycorrhizal Taxa in High-Throughput Sequencing Studies. In: Tedersoo, L. (eds) Biogeography of Mycorrhizal Symbiosis. Ecological Studies 230:125-142. |
|
|
Tedersoo L, Anslan S, Bahram M, Drenkhan R, Pritsch K, Buegger F, Padari A, Hagh-Doust N, Mikryukov V, Gohar D, Amiri R, Hiiesalu I, Lutter R, Rosenvald R, Rähn E, Adamson K, Drenkhan T, Tullus H, Jürimaa K, Sibul I, Otsing E, Põlme S, Metslaid M, Loit K, Agan A, Puusepp R, Varik I, Kõljalg U, Abarenkov K (2020). Regional-Scale In-Depth Analysis of Soil Fungal Diversity Reveals Strong pH and Plant Species Effects in Northern Europe. Frontiers in Microbiology 11:1-31. |
|
|
Tedersoo L, Bahram M, Polme S, Koljalg U, Yorou NS, Wijesundera R, Ruiz L V., Vasco-Palacios AM, Thu PQ, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Rosas M, Riit T, Ratkowsky D, Pritsch K, Poldmaa K, Piepenbring M, Phosri C, Peterson M, Parts K, Partel K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Majuakim L, Lodge DJ, Lee SS, Larsson K-H, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo L -d., Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, De Kesel A, Dang T, Chen X, Buegger F, Brearley FQ, Bonito G, Anslan S, Abell S, Abarenkov K (2014). Global diversity and geography of soil fungi. Science 346:1256688-1256688. |
|
|
Tedersoo L, Jairus T, Horton BM, Abarenkov K, Suvi T, Saar I, Kõljalg U (2008). Strong host preference of ectomycorrhizal fungi in a Tasmanian wet sclerophyll forest as revealed by DNA barcoding and taxon-specific primers. New Phytologist 180:479-490. |
|
|
Thind S, Chaudhary MS, Ditta A, Hussain I, Parveen A, Ullah N, Mahmood Q, Al-ashkar I, El-Sabagh A (2022). Impact of Mycorrhizal Fungi from Different Rhizospheric Soils on Fungal Colonization, Growth, and Chlorophyll Contents of Cenchrus ciliaris. Agronomy 12:2644. |
|
|
Toju H, Sato H, Yamamoto S, Kadowaki K, Tanabe AS, Yazawa S, Nishimura O, Agata K (2013). How are plant and fungal communities linked to each other in belowground ecosystems? A massively parallel pyrosequencing analysis of the association specificity of root-associated fungi and their host plants. Ecology and Evolution 3:3112-3124. |
|
|
Tremblay ÉD, Kimoto T, Bérubé JA, Bilodeau GJ (2019). High-Throughput sequencing to investigate phytopathogenic fungal propagules caught in baited insect traps. Journal of Fungi 5:1-19. |
|
|
Vogelsang KM, Bever JD, Griswold M, Schultz P a (2004). Caltrans Contract The Use of Mycorrhizal Fungi in Erosion Control Applications. |
|
|
van der Wal A, Bloem J, Mulder C, de Boer W (2009). Relative abundance and activity of melanized hyphae in different soil ecosystems. Soil Biology and Biochemistry 41:417-419. |
|
|
Wickham H (2016). ggplot2:Elegant Graphics for Data Analysis. Springer-Verlag New York, Cham. Crossref |
|
|
Wu D, Zhang M, Peng M, Sui X, Li W, Sun G (2019). Variations in soil functional fungal community structure associated with pure and mixed plantations in typical temperate forests of China. Frontiers in Microbiology 10:1-12. |
|
|
Xavier MWF, Rodrigues BF (2020). Identification of Dominant Arbuscular Mycorrhizal Fungi in Different Rice Ecosystems. Agricultural Research 9:46-55. |
|
|
Zhang T, Wang NF, Liu HY, Zhang YQ, Yu LY (2016). Soil pH is a key determinant of soil fungal community composition in the Ny-Ålesund Region, Svalbard (High Arctic). Frontiers in Microbiology 7:1-10. |
|
|
Zhu S, Wang Y, Xu X, Liu T, Wu D, Zheng X, Tang S, Dai Q (2018). Potential use of high-throughput sequencing of soil microbial communities for estimating the adverse effects of continuous cropping on ramie (Boehmeria nivea L. Gaud). PLoS ONE 13:1-16. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0