Full Length Research Paper
ABSTRACT
In Mali, Maure’s zebu breed has good dairy production abilities. Females are often used as a matrix in crosses in peri-urban areas with exotic breeds. Despite the enthusiasm of its productive abilities, the genetic diversity of this breed is not yet sufficiently assessed. In order to contribute to a better understanding of the genetic diversity of this breed, 82 animals were chosen from the farms of four districts in two regions in Mali for genetic evaluation, using Simple Sequence Repeats (SSR) markers. Results from the investigations identified a total of 41 alleles with an average of 3.15 alleles per locus. The Polymorphism Information Content (PIC) ranged from 0.293 (ILSTS011) to 0.786 (HEL5) with a mean of 0.528. By locus, the means of observed heterozygosity (Ho) and expected heterozygosity (He) were 0.400±0,032 and 0.416±0,026, respectively. Likewise, by subpopulation, their values varied respectively from 0.379 ± 0.057 (Kayes) to 0.417 ± 0.070 (Niono) for the observed heterozygosity and from 0.360 ± 0.068 (Baraouéli) to 0.454 ± 0.049 (Dièma) for the expected heterozygosity. The overall mean of genetic differentiation (Fst) was moderate, indicating 9% of the genetic deviation corresponded to differences between subpopulations. This study will strengthen knowledge on the genetic diversity of Maure zebus for the better strategy adoption for its conservation and promotion.
Key words: Maure zebu, genetic, variability, SSR markers, Mali.
INTRODUCTION
Mali has a large herd of cattle in West Africa with the majority concentrated in areas above 600 mm isohyet. Malian cattle breeds are characterized by low production potential (Lhoste, 1983; Nialibouly, 1999), but have on the other hand a great adaptability to the harsh climatic conditions of the Sahel and Sudanian zones. The cattle are comprise zebus (Bos indicus) concentrated in the north and center of the country, taurines (Bos taurus) and the products of zebu-taurine from crosses found in the south of the country (Planchenault, 1988). Among the zebus, the Maure zebu occupies a place of choice among the population who rear it. It is a rather rambling animal, with strong bones (FAO, 1957; Planchenault, 1988). Its limbs are quite strong and well adapted to walking (Planchenault, 1988). The horns are thin, short in the male but longer in the female, they are round in section and greyish or brownish in color (Ministry of Livestock and Fisheries, 2016). The coat is generally dark red, (Planchenault, 1988) but sometimes it could be black and pie-black coats, white on the muzzle on the belly (Ministry of Livestock and Fishing, 2016). The height of the withers varies from 1.40 to 1.50 m for the male and from 1.25 to 1.30 m for the female. The thoracic perimeter varies from 1.60 to 1.85 m with a scapulo-ischial length varying from 1.24 to 1.50 m (Ministry of Livestock and Fisheries, 2016). The weight of the breed is between 250 and 400 kg. Daily milk production varies from 2 to 3 kg reaching 8 kg during good seasons (Nialibouly et al., 2003). Its natural habitat straddles the border between Mali and Mauritania (Ministry of Livestock and Fisheries, 2016). This breed is also known under the following names: Arab zebu, Gabarouyé zebu and Mauritanian zebu (FAO, 1957). In peri-urban rearing, Maure females are increasingly used as a matrix in crosses with exotic breeds for milk production. Good knowledge of the genetic diversity of this breed will enable Malian technicians and breeders to better design genetic improvement and management programs for the Maure zebu. The objective of this study is to assess the genetic diversity of Maure zebu from four districts in two different regions of Mali.
MATERIAL AND METHODS
Ethical considerations
This study titled “Genetic variability study of Maure zebu in Mali using Simple Sequence Repeats (SSR) markers” complies with the Agricultural Orientation Law, the National Policy for Development of Livestock, the National Priority Investment Program for the Agricultural Sector, the Pastoral Charter and the National Strategy for Conservation, Selection and Dissemination of Native Breeds. The study was approved by the ethics committee of the National Institute for Public Health Research under N° 17/2019 / CE-INRSP.
Sampling site
Sampling was carried out in the farms of the Circles of Kayes, Dièma (region of Kayes), Baraouéli and the station of the Regional Agronomic Research Center of Niono (region of Ségou). These sites were chosen according to their accessibility, availability of cow breeders and their herds. The animals were chosen according to the morphological criteria of the breed. A maximum of 5 individuals were sampled per farm. The sites constitute breeding and transit areas for transhumant to the country's wetlands. A sample of 82 heads including 32 from Kayes district, 30 from Dièma district, 8 from Baraouéli district and 12 from Niono district were selected for genetic diversity investigations.
Herd management
The herds were led in an extensive breeding system through the exploitation of natural ranges during the twelve months of the year for the peasant farms of Kayes, Dièma and Baraouéli and in semi-intensive breeding for the animals of the Regional Agronomic Research Center of Niono (CRRA-Niono). Some peasant herds received crop residues and concentrates (cereal bran) during dry periods in addition to grazing. The animals at the Niono CRRA received supplementation (food concentrates), crop residues from rice during the dry season. The health interventions were carried out by the technical breeding agents, if necessary, in the peasant farms. At the Niono CRRA, sanitary prophylaxis was applied to the herd. Mating was natural in all herds. A sire was used for reproduction.
Collection of blood samples
Four milliliters (4 ml) of blood were taken from the jugular vein in EDTA tubes, from males and females randomly selected from the herds. The blood samples were labeled, kept under ice, and transported to the Research Laboratory in Microbiology and Microbial Biotechnology (LaboREM-Biotech) of the Faculty of Sciences and Techniques (FST) of the University of Sciences, Techniques and Technologies of Bamako (USTTB) for molecular analyses.
Extraction of genomic DNA
Zebu genomic DNA samples were extracted from whole blood using the Promega ReliaPrepTM Blood gDNA Miniprep System Extraction Kit. The concentration of the DNAs extracted was determined using the EPPENDORF BioPhotometer® spectrophotometer 6131. DNAs were diluted to 20ng / µl and stored at 4°C for analyzes.
DNA amplification
DNA samples were amplified according to Konaté et al. (2018) with 13 microsatellite primer pairs (Table 1). The markers were chosen from the panel recommended by the International Society of Animal Genetics (ISAG) and the Food and Agriculture Organization (FAO, 2011) for genetic diversity analyzes. These markers have been used in cattle, goat and sheep characterizations (Gororo et al., 2018; Konaté et al., 2018; Al-Atiyat, 2016; Hussain et al., 2016). They are co-dominant, reproducible and polymorphic (Lirón et al., 2002; Portetelle et al., 2000) to generate vital information on the genetic variability of cattle.
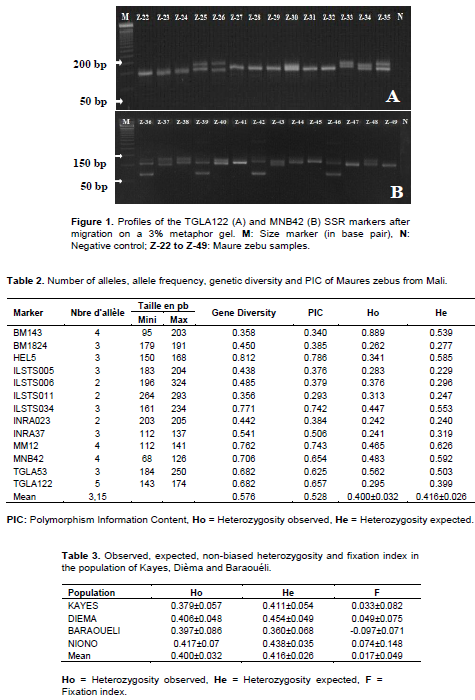
Electrophoresis of amplified products
The gels were prepared with 0.5X TBE (Tris, Borate Acid, EDTA) and 30µl of 10% ethidium bromide (1mg/ml) (Konaté et al., 2018). The MS4 CONDA 3% agarose gel was loaded with 10 μl of each PCR product and migrated for 2 h 30 min at 80V. The gels were photographed with the Gel Documentation System E-BOX VX2 version 15.06.
Statistical analysis
The band (alleles) sizes of each microsatellite primer were determined in base pair using E-Capt software version 15.06. The genetic diversity of each locus was assessed based on the following statistical parameters: number of alleles, frequency of alleles, number of genotypes, genetic diversity, and PIC (Polymorphism Information Content) using Power Marker software version 3.25. The observed (Ho) and theoretical or expected (He) heterozygosity, the overall inbreeding coefficient (Fit), the inbreeding coefficient (Fis), the standardized variance (Fst) were calculated using the GenAIEx software version 6.5. Principal component analysis based on genetic distance was performed with the same software.
RESULTS
Molecular analysis of 82 Maure zebus blood, from four circles (Kayes, Dièma, Baraouéli and Niono) in Mali with thirteen (13) microsatellite markers, revealed a total of 41 alleles forming 73 different genotypes (Figure 1). The number of alleles ranged from 2 (ILSTS006, ILSTS011 and INRA023) to 5 (TGLA122) with an average of 3.15 alleles per locus (Table 2). The PIC ranged from 0.293 (ILSTS011) to 0.786 (HEL5) with a mean of 0.528. Majority of SSR markers exhibited a mean PIC greater than or equal to 0.50. The observed genetic diversity varied from 0.356 to 0.812 respectively for the ILSTS011 and HEL5 loci with an average of 0.576. Values for Ho and He were calculated for each locus and subpopulation under Hardy Weinberg equilibrium. Thus, Ho varied from 0.241 (INRA37) to 0.889 (BM143) with a mean of 0.400±0,032. By subpopulation, it varied from 0.379 ± 0.057 (Kayes) to 0.417 ± 0.07 (Niono) (Table 4). Likewise, He varied from 0.229 (ILSTS005) to 0.626 (MM12) with a mean of 0.416±0,026 (Table 2). Its values per subpopulation from Baraouéli and Dièma varied respectively from 0.360 ± 0.068 to 0.454 ± 0.049 (Table 4).
The markers BM143 and INRA37 loci showed a significant deviation from the Hardy-Weinberg equilibrium in Kayes, Dièma and Niono subpopulations. The MNB42 locus showed a significant deviation from the Hardy-Weinberg equilibrium in Kayes, Dièma and Baraouéli subpopulations. The ILSTS034 locus was significant in Kayes and Dièma subpopulations. Significant deviation from the Hardy-Weinberg equilibrium was noticed with TGLA122 and HEL5 loci in the respective subpopulations of Dièma and Niono (Table 5). The subpopulations of Kayes, Dièma, Baraouéli and Niono presented respective fixation indices F = 0.033 ± 0.082, F = 0.049 ± 0.075, F = -0.097 ± 0.071 and F = 0.074 ± 0.148 with an average of 0.017 ± 0.049. These positive indices reflect a deficit of heterozygosity in the subpopulations, while the negative index indicates an excess of heterozygosity (Table 3).
Wright's F-statistics Fis, Fit and Fst calculated for all Maure zebu populations in the study area were 0.005 ± 0.083, 0.087 ± 0.088 and 0.090 ± 0.022, respectively (Table 5). The overall mean of genetic differentiation (Fst) was moderated indicating that 9% of the genetic deviation corresponded to the differences between populations. All of the amplified microsatellite markers contributed to genetic differentiation (Fst) in sub-populations. However, two microsatellite markers INRA023 (31.3%) and TGLA122 (15.3%) showed significant differentiations. The mean Fis revealed a deficit of heterozygosity in overall population. However, loci BM143, ILSTS005, ILSTS006, ILSTS011, INRA023 and TGLA53 showed excess heterozygosity with negative Fis values (Table 5).
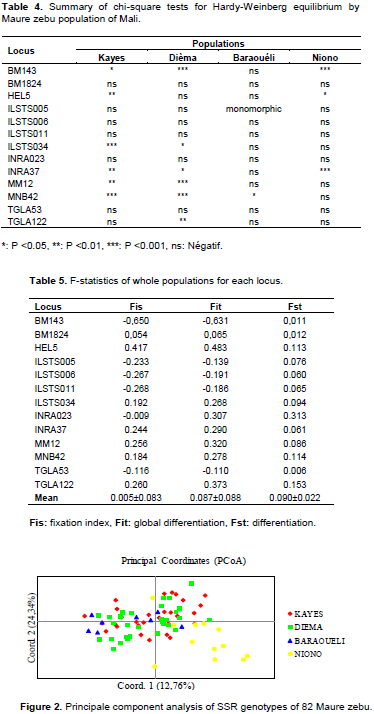
Analysis of Molecular Variance (AMOVA) revealed high genetic variation within individuals (97.33%). The analysis of the principal coordinates revealed a percentage of genetic variation of 12.76 and 24.34% respectively for the first two principal axes with a total of 37.10% (Figure 2). The graph generated by the first two axes, showed individual’s dispersion and do not form specific homogeneous groups. Nevertheless, a mixture of three subpopulations (Kayes, Dièma and Baraouéli) was observed. On the other hand, the subpopulation of Niono was different to that of Kayes, Dièma and Baraouéli.
DISCUSSION
Genetic diversity study of 82 Maure zebus revealed a total of 41 alleles with an average of 3.15 alleles per locus. The mean PIC and the mean genetic diversity were 0.528 and 0.576, respectively. These values ??show a strong diversity within the Maure zebus coming from the various localities of Mali. Study conducted by N’diaye et al. (2015) in Senegal on zebus from Saint-Louis and Kaolack revealed 81 alleles with an average of 7.364 ± 0.544 alleles per locus and an average PIC of 0.719. These values ??are clearly higher than those obtained in the present study and also showed a very high genetic variability of Maure zebus. All the amplified loci were polymorphic based on the mean of the observed PICs. However, 53.85% of the amplified markers showed a PIC greater than or equal to 0.5. On the other hand, this rate is lower than those of N’diaye et al. (2015) on Maure zebus in Senegal, with an average PIC of 100%, greater than 0.5.
The observed mean Ho and expected He recorded at the target loci were 0.400±0,032 and 0.416±0,026, respectively. By subpopulation, He varied from 0.360 ± 0.068 (Baraouéli) to 0.454 ± 0.049 (Dièma) with an average of 0.416 ± 0.026. N’diaye et al. (2015) obtained an average value of 0.769 ± 0.099 on Maure zebus originating from two localities in Senegal. However, in Kayes, Dièma and Niono subpopulations, the observed heterozygosity was less than the expected heterozygosity suggesting a deficit in heterozygosity. Population from Baraouéli showed a great Ho than the He indicating an excess of heterozygosity. The high rate of heterozygosity in this locality could be linked to the contact with other varieties of zebu, the Fulani zebu in particular.
The mean value of the fixation index (Fis) of all the loci was 0.005 ± 0.083, determining a lower heterozygosity deficit compared to the overall population fixation index (Fit = 0.087 ± 0.088). The BM1824, HEL5, ILSTS034, INRA37, MM12, MNB42 and TGLA122 SSR markers contributed to this deficit, through positive Fis values. As for the Fst genetic differentiation, it is relatively very important for the INRA023 locus. In contrast, Fst displays low values ??at the BM143, BM1824, TGLA53 locus levels. The mean differentiation is moderate between subpopulations with a value of 0.090 ± 0.022 indicating the origin of the total genetic variation in the breed.
Principal component analysis (PCA) revealed a mixture of genotypes of the Maure zebu populations of Kayes, Dièma and Baraouéli. Genotypes from Niono were separated from those of other localities. This could be explained by the fact that these animals were bred and selected for several decades for the production of milk. These results reflect the geographic distance between the different districts where the samples were collected. Nearby districts have animals with high genetic similarity. The districts of Kayes and Dièma are contiguous. On the other hand, the district of Niono is more than 700 km away from that of Dièma and Kayes.
CONCLUSION
The molecular characterization of Maure zebu in Kayes, Dièma, Baraouéli and at the station of Niono showed a genetic variability at the loci level. The results from zebu in Kayes and Dièma presented lower observed heterozygosity than the expected one. This supposes that the flock is not subject to genetic introgression from other breeds. In Baraouéli, the observed heterozygosity was higher than the expected one. This can be explained by a possible contact of Maure zebu with Fulani zebu. Baraouéli constitutes the breeding area of Fulani zebu. The obtained results can contribute to build conservations and breeding program of Maure zebu in Dièma, Kayes and Niono.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
ACKNOWLEDGMENTS
The authors appreciate the Competitive Fund for Research and Technological Innovation for its financial support. They are also grateful to the technical livestock services: Local Service for Animal Productions and Industries, Support Units for Animal Productions and Industries, Veterinary Services, as well as the breeders for their support and collaboration.
REFERENCES
|
Al-Atiyat RM (2016). Association of allele diversity and polymorphism of microsatellites markers in the tropical goat. Research Journal of Biotechnology 11(9):29-36. |
|
|
Armstrong E, Postiglioni A, Martínez A, Rincón G, Vega-Pla JL (2006). Microsatellite analysis of a sample of Uruguayan Creole bulls (Bos taurus). Genetics and Molecular Biology 29(2):267-272. |
|
|
Bishop MD, Kappes SM, Keele JW, Stone RT, Sunden SL, Hawkins GA, Yoo J (1994). A genetic linkage map for cattle. Genetics 136(2):619-639. |
|
|
Brzezi?ski T, Majid S (1993). Quantum group gauge theory on quantum spaces. Communications in Mathematical Physics 157(3):591-638. |
|
|
Food and Agriculture Organization (FAO) (1957). African Cattle, Types and Breeds. FAO Agricultural Studie, Rome (37):56-60. |
|
|
Food and Agriculture Organization (FAO) (2011). Molecular genetic characterization of animal genetic resources. FAO Animal Production and Health Guidelines 9 Rome. |
|
|
Georges M, Massey JM (1992). Polymorphic DNA markers in Bovidae. Patent WO 92/13102. |
|
|
Gororo E, Makuza SM, Chatiza FP, Chidzwondo F, Sanyika TW (2018). Genetic diversity in Zimbabwean Sanga cattle breeds using microsatellite markers. South African Journal of Animal Science 48(1):128-141. |
|
|
Hassane YA (2011). Application of microsatellite markers polymorphism for genetic density study of zebus in Niger. Master thesis, University of Polytechnic, Bobo Dioulossao 87, Burkina Faso. |
|
|
Hussain T, Babar ME, Peters SO, Wajid A, Ali A, Azam A, De Donato M (2016). Microsatellite Markers Based Genetic Evaluation of Pakistani Cattle Breeds. Pakistan Journal of Zoology 48(6):1633-1641. |
|
|
Kaukinen J, Varvio SL (1993). Eight polymorphic bovine microsatellites. Animal Genetics 24(2):148. |
|
|
Konaté D, Traoré D, Dao S, Fané R, Ouattara O, Diop R, Babana A (2018). Genetic Diversity Study of N'Dama Breed in Mali Using SSR Markers. International Journal of Animal Science and Technology 2(3):30-35. |
|
|
Lhoste P (1983). Development of animal traction and evolution of pastoral systems in Siné-Saloum, Senegal (1970-1981). Tropical Animal Husbandry and Veterinary Medicine Review 36(3):291-300. |
|
|
Lirón JP, Ripoli MV, De Luca JC, Peral-García P, Giovambattista G (2002). Analysis of genetic diversity and population structure in Argentine and Bolivian Creole cattle using five loci related to milk production. Genetics and Molecular Biology 25(4):413-419. |
|
|
Ministry of Livestock and Fisheries (2016). Directory of cattle, sheep and goat breeds in Mali pp. 31-33. |
|
|
Mommens G, Coppieterst W, Weghe A, Zeveren AV, Bouquet Y (1994). Dinucleotide repeat polymorphism at the bovine MM12E6 and MM8D3 loci. Animal Genetics 25(5):368-368. |
|
|
N'diaye NP, Sow A, Dayo GK, Ndiaye S, Sawadogo GJ, Sembène M (2015). Genetic diversity and phylogenetic relationships in local cattle breeds of Senegal based on autosomal microsatellite markers. Veterinary World 8(8):994-1005. |
|
|
Nialibouly O (1999). Production and reproduction performance of Maure and Fulani zebus (1986-1995) improving at the Niono Agronomic Research Station in Mali: Master thesis, Cheikh Anta Diop University of Dakar, Faculty of Sciences and technologies P 128. |
|
|
Nialibouly O, Coulibaly MD, Dolo D (2003). Improving Dairy Production Performance of Maure and Peul zebu by Selecting at the Niono Station, Mali. Rural Economy Notebooks, Issue (0):3 |
|
|
Planchenault D (1988). L'élevage In : Animal husbandry and sahelian pastoral potentialities. Cartographic synthesis. Mali. CIRAD-IEMVT - FRA. Wageningen: CTA-CIRAD-IEMVT, 26-27. |
|
|
Portetelle D, Haezebroeck V, Mortiaux F, Renaville R (2000). Traçabilité dans la filière animale. Biotechnologie Agronomie Société et Environnement 4(4):233-240. |
|
|
Sodhi M, Mukesh M, Mishra BP, Ahlawat SPS, Prakash B, Sobti RC (2011). Microsatellite analysis of genetic population structure of zebu cattle (Bos indicus) breeds from North-Western region of India. Animal Biotechnology 22(1):16-29. |
|
|
Vaiman D, Mercier D, Moazami-Goudarzi K, Eggen A, Ciampolini R, Lépingle A, Lévéziel H (1994). A set of 99 cattle microsatellites: characterization synteny mapping and polymorphism. Mammalian Genome 5(5):288-297. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0