Full Length Research Paper
ABSTRACT
The widespread emergence of antibiotic resistance, particularly multiple antibiotic resistance (MAR) among bacterial species has become one of the most serious challenges in environmental protection. Environmental bacteria are a reservoir of antibiotic resistance genes (ARGs) and a potential source of novel resistance genes in environmental organisms. In the current study, we investigated the high prevalence of multidrug-resistant Enterobacteriaceae isolated from wastewater and soil in Jos metropolis, Plateau State. A total of 150 wastewater and soil samples were obtained from six different locations within Jos metropolis. Serial dilution was carried out for each sample and inoculated using the spread plate method on Eosin Methylene Blue Agar and MacConkey agar respectively. Total viable count for the environmental isolates was carried out and the isolates were identified macroscopically, microscopically, and biochemically. The antibiotic susceptibility profile of the individual isolates was determined using the Kirby-Bauer disk diffusion method and multiple antibiotics resistance index of the isolates determined. The phenotypic and biochemical identification showed that Escherichia coli has the highest number of occurrences (70%), seconded by Klebsiella spp (20%), and lastly Proteus spp. (10%). It was shown that all the isolates were resistant to Ceftazidime (100%), followed by Ampicillin and Augmentin having (95%) each with Cefuroxime (90%) while Gentamicin has the least resistance with (5%), followed by Ciprofloxacin (15%), Ofloxacin (20%) and Nitrofurantoin (25%). Calculations of MAR for individual bacterial species showed that Klebsiella spp has the highest MAR index of 0.63, followed by E. coli and Proteus spp having MAR index of 0.57, and 0.31 respectively. The study suggests proper management for wastes disposal, the prohibition of unregulated use of antibiotics, and regular monitoring for antibiotics resistance in native bacteria of the environment.
Key words: Antibiotics resistance, public health, MAR Index, environmental waste, enterobacteriaceae.
INTRODUCTION
The appearance of antibiotic resistance poses serious health challenges, economic and social problems because infections caused by antibiotic-resistant bacteria often fail to respond to standard treatments, thereby reducing the possibilities of effective treatment and increasing the risk of morbidity and mortality in serious diseases (Carlet et al., 2011). In the past decades, antibiotic resistance has put increasing pressure globally on human healthcare and is estimated to account for 700,000 deaths every year and the environment has repeatedly been identified as a source for resistant genes to pathogens (Bengtsson-Palme and Larsson, 2016).
One of the most serious challenges in clinical therapy is the widespread emergence of antibiotic resistance, particularly multidrug resistance (MDR), among bacterial pathogens (Levy and Marshall, 2004; World Health Organization, 2000). Acquisition of resistance genes through horizontal transfer is ubiquitous in clinical pathogens (Levy and Marshall, 2004). Environmental bacteria are a reservoir of antibiotic resistance genes and a potential source of novel resistance genes in clinical pathogens (Dantas et al., 2008). Horizontal transfer of genes between bacterial strains could be facilitated by mobile genetic elements, such as plasmids, transposons, bacteriophages, integrons, insertion elements (IS), and genomic islands (Li et al., 2010).
Antibiotic residues contained in the environment are alarming because antibiotics might contribute to the appearance of resistant bacteria and could exert selective pressure. The major source of antibiotics in aquatic environments was once considered to be from hospital sewage, followed by municipal, agricultural, and aquacultural wastewater, which has also been shown to be important sources of these compounds and resistant bacteria (Segura et al., 2009). Treated antibiotic-produced-wastewater contains higher concentrations of antibiotic residues than other aquatic environments, thus can serve as an important reservoir of resistant bacteria and genes (Li et al., 2009, 2008a, b; ?ukasz et al., 2016).
Enterobacteriaceae belongs to a large family of Gram-negative bacteria which are part of the normal gut flora present in the human intestinal tract. Some species can cause diarrhoea and are the common cause of urinary tract infections (UTIs) (Ngene et al., 2020). These pathogens can cause life-threatening complications when they spread to the bloodstream. They include a number of pathogens such as Citrobacter, Salmonella, Klebsiella, Enterobacter, Escherichia coli, Shigella, Proteus, Serratia and other species causing healthcare-associated infections (HAIs). Like all bacteria, enterobacteriaceae can develop resistance to antibiotics which includes the carbapenem group of antibiotics [carbapenem-resistant Enterobacteriaceae (CRE) and carbapenemase-producing Enterobacteriaceae (CPE)] (Yuan et al., 2021).
Among the pathogens disseminated in the environment, enteric pathogens such as enterotoxigenic E. coli, Shigella spp., Salmonella spp., and so forth are the ones most frequently encountered that are responsible for a variety of diseases like diarrhea, dysentery, and enteric fever (Poonia et al., 2014). To further compound this problem, enteric bacterial pathogens have been widely reported to demonstrate resistance to several antibiotics (Chitanand et al., 2010). The environment is the source of bacteria with the highest level of resistance and surface water is the main reservoir of antibiotics and antibiotic-resistant bacteria in the environment. In the past two decades, the rise in antibiotic resistance has been reported and remains a global problem (Sharma and Rai, 2012; Verma et al., 2011). In the current study, we investigated the high prevalence of multidrug-resistant Enterobacteriaceae isolated from wastewater and soil in Jos Metropolis, Plateau State.
MATERIALS AND METHODS
Collection of samples
A total of 150 wastewater and soil samples (25 samples for each location) were obtained from 6 different locations (Student Village Hostels 1 and 2, Old Jos University Teaching Hospital, JUTH 1 and 2, and Angwa Rukuba 1 and 2), within Jos North Metropolis, Plateau State, Nigeria. Latitude and Longitudes of their various locations were noted. A 50-ml sterile vial with cover tops were used for this purpose. The containers were immediately disinfected with 70% ethanol at the point of collection, labeled, and kept in a super cool flask for transportation to Africa Center of Excellence in Phytomedicine Research and Development, ACEPRD, University of Jos, Microbiology Laboratory for analysis.
Laboratory Isolation
According to the modified method cited by Ibrahim and Hameed ( 2015), a total of 10 ml of each sample (after mixing the wastewater and sand and allowed to decant in a conical flask) was diluted in 90-ml of sterile 0.9% NaCl normal saline and homogenized. Thereafter, 100 μl of the fourth and fifth diluent of the samples were inoculated on Eosin Methylene Blue Agar (EMB) agar plates for the isolation of enteric bacteria and MacConkey agar plates are used for both lactose and non-lactose fermenters bacterial isolates using the spread plate method. All the bacteria plates were incubated at 37?C for 24 h.
Total viable count for environmental isolates
The total viable count was determined using the spread plate technique on nutrient agar and counting the colonies developed after incubation at 37?C for 24 h (Harley and Prescott, 1996).
Identification of isolates
Gram-negative bacteria were isolated on their respective selective and differential media and were identified based on culture characteristics, including Gram stain, motility, and biochemical tests, MacConkey agar, EMB, IMViC, urea, and triple sugar iron (TSI) test (Forbes et al., 2016).
Preservation of isolates
The isolates were subcultured on nutrient agar, incubated at 37°C for 24 h. A single colony was inoculated into a sterile nutrient broth, incubated in a shaker incubator (ZHP-100) at 180 rpm for 24 h at 37°C. The isolates were also incubated on a nutrient agar slant at 37°C for 24 h. They were all stored at 4°C in a refrigerator.
Antibiotics susceptibility profile
The antibiotic susceptibility profile of the Gram-negative isolates was determined using the standard Kirby-Bauer disk diffusion method (Bauer, 1966). These antibiotics with their respective disk concentrations are as follows: Ceftazidime (10 μg), Cefuroxime (30 μg), Gentamicin (10 μg), Ciprofloxacin (10 μg), Nitrofurantoin (300 μg), Ampicillin (10 μg), Ofloxacin (10 μg), and Augmentin (30 μg) (Bhattacharya et al., 2012). Bacterial culture suspension equivalents of 0.5 tube McFarland turbidity standards were spread on Muller-Hinton agar plates using sterile swabs and incubated aerobically at 37?C for 24 h; then, the diameters of the zone of inhibition surrounding the antibiotic disks was measured. The results are expressed as susceptible or resistant according to the criteria recommended by (CLSI, 2012).
Multiple antibiotics resistance (MAR) index
This MAR index was suggested by Krumperman (1983), according to the following formula in Equation 1 and 2.
MAR = a/b (1)
Where; a = the number of antibiotics to which the isolate was resistant; b = the number of antibiotics to which the isolate was exposed.
MAR = a/(b × c) (2)
Where; a = the aggregate antibiotic resistance score of all isolates from the sample; b = the number of antibiotics; c = the number of isolates from the sample. Also, values of MAR greater than 0.25 pose a high-risk source for contamination.
Statistical analysis
All the experiments were repeated three times and the mean values of the three replicates obtained. The statistical analysis was carried out using SPSS software version 21. Data were analyzed to determine the analysis of variance (ANOVA) using Duncan’s multiple range test (JMP v.12 software; SAS Inst., Cary, NC, USA). Significant differences between results were estimated at a P-value less than 0.05 (P < 0.05).
RESULTS
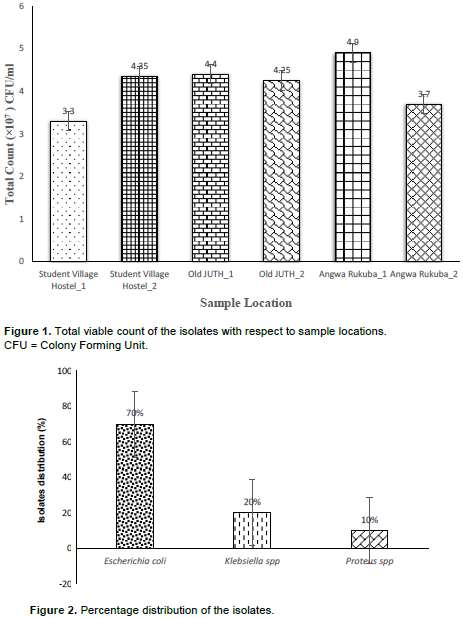
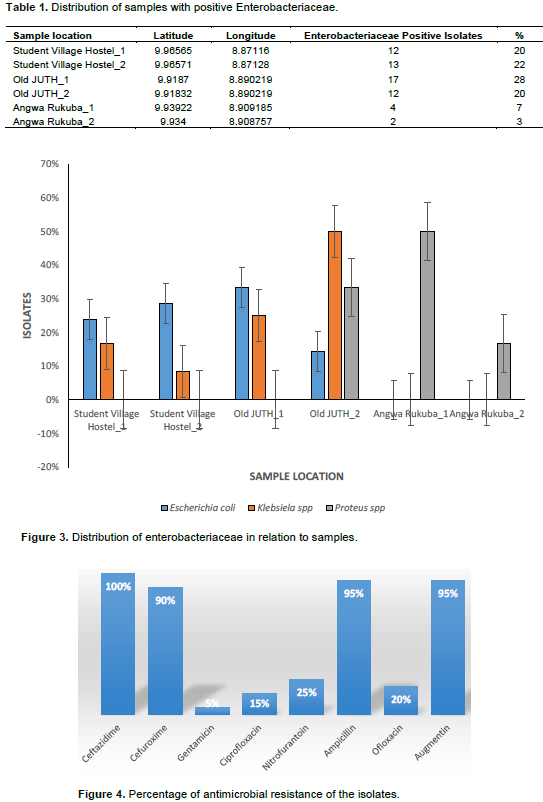
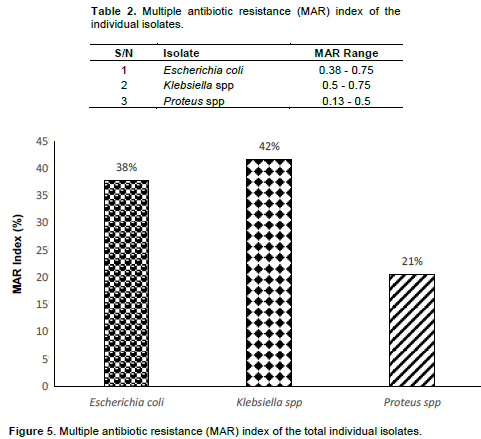
In the present study, the samples were collected in six different locations within the Jos metropolis. Angwa Rukuba_1 having the highest mean value of total viable bacteria count (4.9x107 CFU/ml), followed by Old JUTH_1 (4.4x107 CFU/ml), Student Village Hostel_2 (4.35x107 CFU/ml), Old JUTH_2 (4.25x107 CFU/ml), Angwa Rukuba_2 (3.7x107 CFU/ml) and Student Village Hostel_1 having the least viable count (3.3x107 CFU/ml) as shown in Figure 1. As illustrated in Figure 2, the phenotypic and biochemical identification showed that E. coli has the highest number of occurrences (70%), seconded by Klebsiella spp (20%) and lastly Proteus spp. (10%). Table 1 showed that Old JUTH 1 has the highest number of positive Enterobacteriaceae (28%), followed by Student Village Hostel_2 (22%). Student Village Hostel_1 and Old JUTH 2 have the same number of Enterobacteriaceae (20%) each while Angwa Rukuba 1 and 2 had the least 7 and 3% respectively. Figure 3 showed that Old JUTH_1 (33%) has the highest distribution of E. coli to sample location, followed by Student Village Hostel_2 (29%), Student Village Hostel_1 (24%), and Old JUTH_2 having the least (14%) while Angwa Rukuba 1 and 2 recorded none. For Klebsiella spp., Old JUTH_2 had the highest distribution number of (50%), followed by Old JUTH_1 (25%), Student Village Hostel_1 (17%), and the least Student Village Hostel_2 with (8%) and was absent in Angwa Rukuba 1 and 2. Angwa Rukuba_1 has the highest distribution number of Proteus spp. (50%) followed by Old JUTH_2 (33%) and the least Angwa Rukuba_2 (17%). As demonstrated in Figure 4, it was shown that all the isolates were resistant to Ceftazidime (100%), followed by Ampicillin and Augmentin having (95%) each with Cefuroxime having (90%). Gentamicin had the least resistance with (5%), followed by Ciprofloxacin, Ofloxacin, and Nitrofurantoin having 15, 20, and 25% respectively. Susceptibility of bacteria to different antibiotics (8 items) showed multiple antibiotics resistance (MAR) for the majority of isolates. As indicated in Table 2 and illustrated by Figure 5, calculations of MAR for individual bacterial species showed that Klebsiella spp has the highest MAR index of 0.63, followed by E. coli and Proteus spp having MAR index of 0.57, and 0.31 respectively.
DISCUSSION
There is a need for periodic surveillance of laboratory activities to monitor antibiotic resistance and its spread in our environment. This will help in gathering information needed in making policies that matter on antimicrobial resistance (World Health Organization, 2013). It is worth mentioning that, all the study samples exceeded the international standard limits (5000 CFU 100 ml−1) (Collivignarelli et al., 2017; Tebbutt, 1998) and could be a result of fecal contamination as reported by Azzam et al. (2017). Some restricted limits have been reported by Efstratiou et al. (2009) and Cabelli (1978), a maximum total coliforms count of 1000 CFU 100 ml−1, particularly in surface water that would be used as a drinking water supply. Bacteria generally identified in this study were reported to be potential human pathogens of a public health concern as described by Sneath (1986), Cheesbrough (2006), and World Health Organization (2011). The most widespread bacteria obtained was E. coli, followed by Klebsiella spp and Proteus spp which indicates that the samples were subjected mainly to sewage pollution as reported by Ibrahim and Hameed (2015) which recorded E. coli to be the most common lactose-fermenting bacterial isolates from the environmental specimens, comprising 54.6% of the total samples, followed by Klebsiella pneumonia with 32.8% of samples. The high incidence of E. coli correlated with fecal coliforms supports such findings (Edberg et al., 2000; Azzam et al., 2017). The environmental isolated Enterobacteriaceae showed a high level of resistance to Ceftazidime, Cefuroxime, Ampicillin, and Augmentin while susceptible to Gentamicin, Ciprofloxacin, Ofloxacin, and Augmentin which supports the research findings of Ibrahim and Hameed (2015) and Azzam et al. (2017). The high susceptibility profile of the bacterial isolates to the named antibiotics could be related to the less frequent use of these drugs for therapeutic purposes, therefore reducing the chance for resistance as reported by Ibrahim and Hameed (2015). The genetic background of resistance mechanisms is diverse because they are present on chromosomes, plasmids, integrons, and transposons (Brooks et al., 2010). High levels of genetic flux between Gram-negative Enterobacteriaceae have been suggested by many studies (Stecher et al., 2012) that may favor exchange of plasmid between Enterobacteriaceae members. Study area with MAR index values above 0.25 as calculated by Krumperman (1983) and Hinton et al. (1985) was classified as potential health risk environments. In both drainage and river water, Munir et al. (2011) reported in Michigan (USA) the incidence of MAR bacteria and was also reported in the work of Azzam et al. (2017) in Egypt. This shows that the issue of multiple antibiotics resistant bacteria in the environment is of global concern since it is of international, rather than national problem (Knapp et al., 2012; Lupan et al., 2017; Okeke and Edelman, 2001).
CONCLUSION
The study shows that the Enterobacteriaceae isolated were E. coli, Klebsiella spp, and Proteus spp, which demonstrated multidrug resistance for Ceftazidime, Cefuroxime, Ampicillin, and Augmentin. Factors that may be associated with the transmission of resistant strains in the environment include poor hygiene and antibiotic abuse. More bacterial isolates from different sources in conjunction with genetic analysis are to be collected for future studies.
This situation suggests regular monitoring for antibiotics resistance in native bacteria of the environment, the prohibition of unregulated use of antibiotics, and proper management for wastes disposal.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
ACKNOWLEDGEMENTS
The authors express their gratitude to the Director, Africa Center of Excellence in Phytomedicine Research and Development University of Jos, Plateau State, Nigeria for funding this research. World Bank also sponsored and is appreciated.
REFERENCES
|
Azzam MI, Ezzat SM, Othman BA, El-Dougdoug KA (2017). Antibiotics resistance phenomenon and virulence ability in bacteria from water environment. Water Science 31(2):109-121. |
|
|
Bauer AW (1966). Antibiotic susceptibility testing by a standardized single disc method. American Journal of Clinical Pathology 45:149-158. |
|
|
Bengtsson-Palme J, Larsson DJ (2016). Concentrations of antibiotics predicted to select for resistant bacteria: proposed limits for environmental regulation. Environment international 86:140-149. |
|
|
Bhattacharya D, Sugunan AP, Bhattacharjee H, Thamizhmani R, Sayi DS, Thanasekaran K, Roy S (2012). Antimicrobial resistance in Shigella-rapid increase & widening of spectrum in Andaman Islands, India. The Indian Journal of Medical Research 135(3):365. |
|
|
Brooks GF, Carroll KC, Butel JS, Morse SA, Mietzner TA (2010). Medical Microbiology. Jawetz, Melnick and Adelbergs, 25th Edition, McGraw-Hill Companies pp. 213-219. |
|
|
Cabelli V (1978). New standards for enteric bacteria. Water Pollution Microbiology 2:233-273. |
|
|
Carlet J, Collignon P, Goldmann D, Goossens H, Gyssens IC, Harbarth S, Richtmann R (2011). Society's failure to protect a precious resource: antibiotics. The Lancet 378(9788):369-371. |
|
|
Cheesbrough M (2006). District laboratory practice in tropical countries, part 2. Cambridge University Press. |
|
|
Chitanand MP, Kadam TA, Gyananath G, Totewad ND, Balhal DK (2010). Multiple antibiotic resistance indexing of coliforms to identify high risk contamination sites in aquatic environment. Indian Journal of Microbiology 50(2):216-220. |
|
|
Clinical and Laboratory Standards Institute (CLSI) (2012). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement. CLSI Document M 100-S22. Clinical and Laboratory Standards Institute, Wayne. |
|
|
Collivignarelli MC, Abbà A, Alloisio G, Gozio E, Benigna I (2017). Disinfection in wastewater treatment plants: evaluation of effectiveness and acute toxicity effects. Sustainability 9(10):1704. |
|
|
Dantas G, Sommer MOA, Oluwasegun RD, Church GM (2008). Bacteria subsisting on antibiotics. Science 320(5872):100-103. |
|
|
Edberg SCL, Rice EW, Karlin RJ, Allen MJ (2000). Escherichia coli: the best biological drinking water indicator for public health protection. Journal of Applied Microbiology 88(S1):106S-116S. |
|
|
Efstratiou MA, Mavridou A, Richardson C (2009). Prediction of Salmonella in seawater by total and faecal coliforms and Enterococci. Marine Pollution Bulletin 58(2):201-205. |
|
|
Forbes BA, Sahm DF, Weissfeld AS (2016). Study Guide for Bailey and Scott's Diagnostic Microbiology-E-Book. Elsevier Health Sciences. |
|
|
Harley JP, Prescott LM (1996). Microbiology: Laboratory Exercises. 3rd Edition, McGraw-Hill Companies, New York. |
|
|
Hinton M, Hedges AJ, Linton AH (1985). The ecology of Escherichia coli in market calves fed a milk?substitute diet. Journal of Applied Bacteriology 58(1):27-35. |
|
|
Ibrahim IAJ, Hameed TAK (2015). Isolation, characterization and antimicrobial resistance patterns of lactose-fermenter enterobacteriaceae isolates from clinical and environmental samples. Open Journal of Medical Microbiology 5(4):169-176. |
|
|
Knapp CW, Lima L, Olivares-Rieumont S, Bowen E, Werner D, Graham DW (2012). Seasonal variations in antibiotic resistance gene transport in the Almendares River, Havana, Cuba. Frontiers in Microbiology 3:396. |
|
|
Krumperman PH (1983). Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Applied and Environmental Microbiology 46(1):165-170. |
|
|
Levy SB, Marshall B (2004). Antibacterial resistance worldwide: causes, challenges and responses. Nature Medicine 10(12):S122-S129. |
|
|
Li D, Yang M, Hu J, Ren L, Zhang Y, Li K (2008a). Determination and fate of oxytetracycline and related compounds in oxytetracycline production wastewater and the receiving river. Environmental Toxicology and Chemistry: An International Journal 27(1):80-86. |
|
|
Li D, Yang M, Hu J, Zhang J, Liu R, Gu X, Wang Z (2009). Antibiotic?resistance profile in environmental bacteria isolated from penicillin production wastewater treatment plant and the receiving river. Environmental Microbiology 11(6):1506-1517. |
|
|
Li D, Yang M, Hu J, Zhang Y, Chang H, Jin F (2008b). Determination of penicillin G and its degradation products in a penicillin production wastewater treatment plant and the receiving river. Water Research 42(1-2):307-317. |
|
|
Li D, Yu T, Zhang Y, Yang M, Li Z, Liu M, Qi R (2010). Antibiotic resistance characteristics of environmental bacteria from an oxytetracycline production wastewater treatment plant and the receiving river. Applied and Environmental Microbiology 76(11):3444-3451. |
|
|
?ukasz J, Joanna C, P?aza G, Dorgeloh E, Ejhed H (2016). Antibiotic susceptibility of bacteria isolated from onsite wastewater treatment facilities. International Multidisciplinary Scientific GeoConference: SGEM 1:397-404. |
|
|
Lupan I, Carpa R, Oltean A, Kelemen BS, Popescu O (2017). Release of antibiotic resistant bacteria by a waste treatment plant from Romania. Microbes and Environments 32(3):219-225. |
|
|
Munir M, Wong K, Xagoraraki I (2011). Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Research 45(2):681-693. |
|
|
Ngene AC, Aguiyi JC, Uzal U, Egbere JO, Onyimba IA, Umera AE, Nnadi NE (2020). Bacteriophages as Bio-control agent against Food-Borne Pathogen E. coli O157: H7. IOSR Journal of Pharmacy and Biological Sciences 15(2):23-36. |
|
|
Okeke IN, Edelman R (2001). Dissemination of antibiotic-resistant bacteria across geographic borders. Clinical Infectious Diseases 33(3):364-369. |
|
|
Poonia S, Singh TS, Tsering DC (2014). Antibiotic susceptibility profile of bacteria isolated from natural sources of water from rural areas of East Sikkim. Indian journal of community medicine: official publication of Indian Association of Preventive & Social Medicine 39(3):156. |
|
|
Segura PA, François M, Gagnon C, Sauvé S (2009). Review of the occurrence of anti-infectives in contaminated wastewaters and natural and drinking waters. Environmental Health Perspectives 117(5):675-684. |
|
|
Sharma BC, Rai B (2012). Incidence of multi-drug resistance in E. coli strains isolated from three lakes of tourist attraction (Mirik lake, Jorepokhani lake and Nakhapani lake) of Darjeeling Hills, India. Indian Journal of Fundamental and Applied Life Science 2:108-14. |
|
|
Sneath PH (1986). Bergey's Manual of Systematic Bacteriology, Vol. 2. Williams and Wilkins Baltimore, London, Los Angeles, Sydney, USA. |
|
|
Stecher B, Denzler R, Maier L, Bernet F, Sanders MJ, Pickard DJ, Ackermann M (2012). Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proceedings of the National Academy of Sciences 109(4):1269-1274. |
|
|
Tebbutt T (1998). Principles of Water Quality Control, 5th ed. Hallam University. |
|
|
Verma NS, Gupta A, Dubey M, Mahajan S, Sharma R (2011). Resistance status of some pathogenic bacteria isolated from water of Yamuna river in Agra. Asian Journal of Experimental Biological Sciences 2:697-703. |
|
|
World Health Organization (WHO) (2000). World Health Organization report on infectious diseases 2000-overcoming antibiotic resistance. World Health Organization, Geneva, Switzerland. |
|
|
World Health Organization (WHO) (2011). Guidelines for Drinking-Water Quality, 4th ed. WHO, Geneva. |
|
|
World Health Organization (WHO) (2013). Antimicrobial Resistance: Global Report on Surveillance, Geneva. |
|
|
Yuan W, Zhang Y, Riaz L, Yang Q, Du B, Wang R (2021). Multiple antibiotic resistance and DNA methylation in Enterobacteriaceae isolates from different environments. Journal of Hazardous Materials 402:123822. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0