ABSTRACT
Escherichia coli O157:H7 is among the most pathogenic of all known foodborne pathogens. It causes severe diarrhoea with apparently low infective dose (< 10 cells). This study aimed to determine the prevalence of E. coli O157:H7 in foods of animal sources sold in Sokoto Metropolis, Sokoto, Nigeria. A total of 175 samples were collected from different locations within Sokoto metropolis. Culture and biochemical characterisation revealed E. coli with an overall detection rate of 50.9% (89/175) with percentages of isolation rates of 30% (12/40), 75% (30/40), 43.6% (24/55) and 57.5% (23/40) for fresh milk, fermented milk, egg and raw meat respectively. Further characterization of the isolated E. coli on Sorbitol MarcConkey (SMAC) agar yielded E. coli O157:H7 strain with a positive detection rate of 31.4% (55/175) comprising 22.5% (9/40), 50.0% (20/40), 18.2% (10/55) and 40.0% (16/40) for fresh milk, fermented milk, egg and raw meat respectively. Molecular identification of shiga-toxin 1 (Stx I) and shiga-toxin 2(Stx II) genes in the E. coli O157:H7 isolates by polymerase chain reaction (PCR) yielded 10 amplicons of Stx 1 genes and 6 amplicons of Stx II genes. The study confirmed the presence of toxigenic E. coli O157:H7 in animal products sold within Sokoto metropolis. The application of Hazard Analysis Critical Control Point (HACCP) protocol in the production processes is recommended to identify probable sources of microbial contaminants and to appropriately prevent contamination. The public should be enlightened on the zoonotic potential of this foodborne pathogen and the role of good hygiene practices in food safety.
Keywords: Escherichia coli O157:H7, Shiga-like toxin, Sorbitol MacConkey agar.
Escherichia coli are Gram-negative, facultative anaerobic bacteria that belong to the family Enterobacteriaceae. The bacterium is typically rod-shaped and about 2 µm long and 0.5 µm in diameter. Some strains possess flagella which enable the bacterium to move (Xia et al., 2010). The E. coli strains causing enteric diseases are categorized by their symptoms, virulence-factors, and the pathomechanisms that led to their categorization into various pathotypes such as enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli (EHEC), verotoxin-producing E. coli (VTEC), enteroaggregative E. coli (EAggEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC) and the diffusely adherent E. coli (DAEC). E. coli strains responsible for urogenital infections (UPEC), and sepsis or meningitis of the neonates (MAEC) belong to the group causing extra-intestinal outbreaks. These pathogens are transmitted to humans through consumption of contaminated foods, such as raw or undercooked vegetables, meat, milk and their products (Castro et al., 2017). The most important common property of the pathogens is the possession of virulence-factors that are encoded on a variety of mobile genetic elements such as on plasmids, bacteriophages, transposons and pathogenicity islands. The harboured adhesins and toxins enable the colonisation of the intestinal mucosa, differently from the non-pathogenic members of the normal intestinal flora, thus their ability to cause a wide range of enteric infections (Schaeffer, 2012).
The first reported case of E. coli O157:H7 haemorrhagic colitis was in 1990. Since then, many sporadic cases of bloody diarrhoea have been reported in many areas of South Africa. Effer et al. (2001) reported verotoxigenic E. coli from South Africa in 1992, a large outbreak of bloody diarrhoea caused by E. coli O157:H7 infections occurred in Swaziland, SouthAfrica. About 40,912 patients were suspected to be infected. The attack rate was 42% among 778 residents screened. Female gender and consumption of beef and untreated water were significant risks for the illness. E. coli O157:H7 was recovered from seven affected foci in Swaziland and South Africa, 27 out of 31 patients and environmental isolates had undistinguished pulsed field gel electrophoresis patterns. Cattle deaths also occurred due to verotoxigenic E. coli. Drought carriage of E. coli O157:H7 by cattle and heavy rains with contamination of surface water appear to be important factors contributing to verotoxigenic outbreaks. Molecular techniques were also used for studying the epidemiology of diaarhea infections due to E. coli in Gauteng region of South Africa.
In Nigeria, Akinyemi et al. (1998), studied E. coli infections for over 12 months. A total of 852 stool samples from patients (both children and adults) with acute diarrhoeal diseases attending some public and some government recognised health institutions in Lagos metropolis were screened for diarrhoeagenic bacterial agents. Of all 83 isolates for E. coli group,49(59%) were EPEC ,17(20.5%), ETEC, 10(12.1%) EIEC and seven (8.4%) EHEC. The EPEC strains particularly serotype 055, were mostly encountered in children aged over five years. On the other hand, EIEC and ETEC strains were found mainly in adults while EHEC O157:H7 strains occurred in all age group studied (Akinyemi et al., 1998). Olorunshola et al, (2000) examined the prevalence of sorbitol non- fermenting E. coli O157:H7(EHEC) in 100 patients with diarrhoea by stool culture on sorbitol mac Conkey agar in Lagos, Nigeria. The authors reported 6% detection rate of E. coli O157:H7 and five of the six patients were children below five years of age and a teenager. This study was aimed at determining the prevalence of E. coli O157:H7 in milk, egg and meat sold within Sokoto Metropolis, Sokoto, Nigeria.
Study area
The study was conducted in Sokoto metropolis. The State lies within the semi-arid region of north-western Nigeria between longitudes 4°8`E and 6°54`E and latitudes 12°N and 13°58`N. It covers a total land area of about 32,000 square km. The estimated population of the State as at 2016 is about 5 million (NBS, 2017) with an estimated animal population of 1.8 million cattle, 2.6 million sheep, 2.9 millon goats 48,000 camels and variable species of poultry (RIMS, 1991; MAHF, 2012).
Sample collection
A total of 175 samples comprised up of 40 samples each of fresh milk, fermented milk and raw meat and 55 eggs from different outlets within Sokoto metropolis were collected. Fresh and fermented milk samples were aseptically collected in a sterile screw-capped bottle from identified dairy farms within the metropolis and central diary outlet popularly known as ‘Kasuwar Gawo’ respectively. Similarly, raw meat and eggs samples were aseptically collected from different outlets into a sterile polythene bag and in a clean crate respectively. All the samples collected were immediately transported in an insulating flask with ice packs to the Veterinary Microbiology laboratory for analyses.
Culture, isolation and identification
All the samples collected were pre-enriched in buffered peptone broth. A gram of meat samples (already cut into small portions with a sterile blade) and 1 ml of both fresh and fermented milk samples and egg wash samples were pre-enriched by inoculating into 9mls of buffered-peptone broth each in different test tubes, homogenized and incubated at 37°C for 24 h. After incubation, a loopful inoculum from peptone broth was streaked onto MacConkey and Eosin Methylene Blue (EMB) agar plates and incubated at 37°C for 24 h. Those pinkish colonies on MacConkey and greenish metallic sheen appearance on EMB agar were presumptively identified as E. coli and thus, selected and subcultured for further phenotypic and biochemical analysis; these include Gram staining, Indole, methyl red (MR), Voges Proskauer (VP) and Citrate tests. Those Gram negative isolates that are Indole and MR tests Positive with VP and Citrate tests negative were confirmed as E. coli. Confirmed E. coli isolates were further characterized on Sorbitol macConkey agar for identification of E. coli O157:H7 from E. coli non-O157:H7 strains. Smooth and colourless colonies (Non-sorbitol fermenters) were phenotypically identified as E. coli O157:H7 as illustrated elsewhere (Safarikova and Safarik, 2001; Atikson et al., 2012).
DNA extraction
The genomic DNA of identified E. coli O157:H7 isolates were extracted using boiling method as described elsewhere (Junior et al., 2016). Briefly, a loopful of the 18-24 h old E. coli O157:H7 isolates was suspended in 200 µl of molecular-grade water in a microcentrifuge tubes. The suspension was heated in a water bath at 96°C for 30 min and centrifuged at 1300 rpm for 2 min. The supernatant (DNA templates in solution) was used as DNA template in polymerase chain reaction (PCR) techniques.
Detection OF E. coli O157:H7 virulence genes (STX I & STX II)
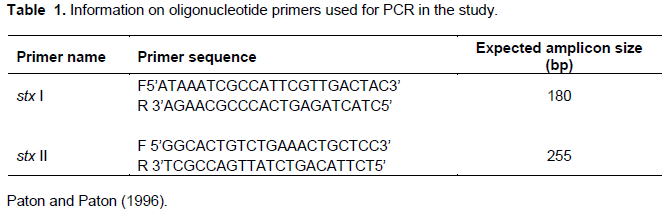
A multiplex PCR was conducted using TopTaq™ Master Mix PCR kit (Biolabs®) using extracted genomic DNA of E. coli O157:H7 isolates. The PCR was conducted with 25 µl reaction mixture containing TopTaq™ Master Mix (12.5 µl), RNase-free water (7.5 µl), DNA template 200 ng (2.5 µl) and 1 µl of four-primer cocktail (0.25 µM) (nucleotide sequence in Table 1). The primers are amplifying Stx1 (180 bp) and Stx2 (255 bp) genes respectively as adopted (Paton and Paton, 1996). Amplification was conducted in Geneamp 9700 PCR system (Applied Biosystem). The reaction mixtures were subjected to cycling parameters of 35 cycles of 1 min of denaturation at 95°C, 2 min of annealing at 65°C for the first 10 cycles, decrementing to 60°C by cycle 15, and 1.5 min of elongation at 72°C, incrementing to 2.5 min from cycles 25 to 35. Template DNA of a confirmed E. coli O157:H7 and sterile molecular-grade water were used as positive and negative controls respectively. Before loading samples into agarose-gel wells, 2 µl of DNA ladder was mixed with 2 µl of loading dye and dispensed in the first well. Subsequent wells were loaded with 5 µl samples of the PCR product and analyzed using 1.5% agarose gel electrophoresis and viewed in a documentation system (Gel Doc™ XR+, Bio- Rad).
Data analysis
The data were presented in tables and charts. Descriptive statistics were used to display the distribution of shiga-toxin 1 and 2 genes in the E. coli isolates.
Out of the total (n=175) samples collected; which comprises fresh milk (n=40), fermented milk (n=40), egg (n=55) and raw meat (n=40). The overall prevalence of E. coli was 50.9 (89/135) with percentages isolation rates of 30% (12/40), 75% (30/40), 43.6% (24/55) and 57.5% (23/40) for fresh milk, fermented milk, egg and raw meat respectively (Table 2).
Out of total 89 E. coli isolates, 55 were identified as E. coli O157:H7 with an overall detection rate of 31.4% (55/175) which comprises of 22.5% (9/40), 50.0% (20/40), 18.2% (10/55) and 40.0% (16/40) for fresh milk, fermented milk, egg and raw meat respectively (Figure 1). DNA templates of the fifty-five E. coli O157:H7 isolates (previously isolated from food samples) were subjected to a polymerase chain reaction in search of shiga-toxin1 (Stx I) and shiga-toxin 2 (Stx II) genes which indicate Shiga-like toxin-producing E. coli O157:H7. The result of this study yielded 10 amplicons of Stx 1 genes and 6 amplicons of Stx II genes (Figure 2). The overall prevalence of Shiga-like toxin-producing E. coli O157:H7 stood at 5.7% (10/175) which comprises of 2.3% (4/175) those harbouring Stx I gene alone and 3.4% (6/175) harbouring Stx I and II together. This indicated that the remaining 94.3% (165/175) are non-toxigenic E. coli O157:H7 (Table 3). Out of the nine E. coli O157:H7 isolates obtained from fresh milk samples, only one isolate is positive (and both stx I and stx II genes were amplified). Similarly, out of twenty E. coli O157:H7 isolates obtained from fermented milk samples, three showed positives for StxI among which one (lane 5, Figure 2) showed both amplification for StxI and Stx II genes. Of the ten (10) E. coli O157:H7 isolates obtained from egg samples, five showed positive for StxI gene among which four showed positive for both Stx 1 and StxII genes Lastly, out of the sixteen E. coli O157:H7 isolates gotten from meat samples, only one showed the presence of StxI gene with no amplification in Stx II gene (Figure 2).

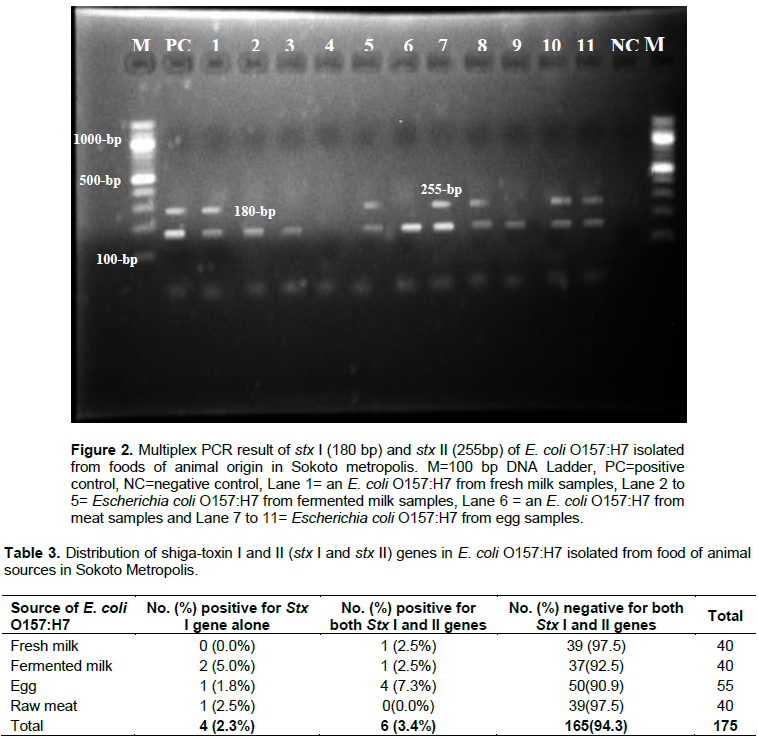
E. coli, especially Shiga-toxin producing strains, are an important cause of diarrhoea and gastrointestinal illness in humans and animals especially young. Some of which are life-threatening such as haemolytic-uremic syndrome (an important cause of acute renal failure in children with morbidity and mortality in adults) and haemorrhagic colitis and thrombotic thrombocytopenic purpura (Al-Zogibi et al., 2015). Fresh and fermented milk is known to be widely consumed in both rural and urban areas in the study area. This might be due to its affordability and availability. Fermented milk is obtained from fresh milk that had undergone series of processing before finally converted to fermented milk, however, several cross contaminations do occur during collection (unclean hands of worker, unhygienic condition of utensils, and unclean water used for washing the utensils), handling, processing, transportation and marketing, therefore, exposes human population at risk of getting E. coli infection. Of economic importance, however, is the occurrence of pathogenic strains of E. coli O157:H7 in milk, meat and egg samples analysed in this study, which could be hazardous to consumers. The prevalence of E. coli O157:H7 in milk and milk products was found to vary between 1.0 and 11.0% (Reuben et al., 2002; Yakubu et al., 2018). The survival of this pathogen in low pH milk derivatives has also been documented in the various literatures (Reuben et al., 2002).
The method of handing, transporting and marketing of the raw meat and eggs are unhygienic. Similarly, the raw meat and eggs fall on an easy prey to bacterial contamination because of the high ambient temperature of Sokoto state. Such condition could pose favorable environment for bacterial contamination of the product. Raw beef, vegetables and milk products have been described as the principal vehicle of E. coli O157:H7 transmission to humans (Reuben et al., 2002; Castro et al., 2017; IFSAC, 2019). Global testing of beef had shown E. coli O157:H7 prevalence that ranges between 0.1 and 54.0% (Chapman et al., 2001; Hussein and Bollinger, 2005) and had been isolated from retail meat samples in many developing countries such as Thailand (Vuddhakul et al., 2000), South Africa (Mukhufhi et al., 2004), Saudi Arabia (Al-Humam, 2019) and in Algeria (Chahed et al., 2006), with prevalence that ranges between 4 and 9%.
CONCLUSION AND RECOMMENDATIONS
The results obtained from this study confirmed the presence of E. coli O157:H7 in some food items of animal origin (milk, eggs and meat) traded for human consumption in Sokoto metropolis. The presence of both Stx 1 or Stx I and Stx II affirmed the virulence of the E. coli O157:H7 strains isolated in this study. The prevalence of Stx I (10 amplicons) was found to be more as compared to Stx II (6 amplicons) which is a peculiar characteristic of the genes when amplified. The milk, eggs and meat could be contaminated with these pathogens along the production line, during storage or in the course of transportation. This situation highlights a serious concern and threat to public health as naïve and less immunocompetent hosts (young, elderly and immunodeficient individuals) may fall prey to this pathogens. The research findings pointed at the need for total overhaul of the existing methods of milk production in the study area. This may include the application of hazard analysis critical control point (HACCP) to guide the identification of probable sources of contaminations in order to ascertain, mitigate and outline prevention measures. Primary health workers could be trained on hazard analysis critical control to ensure improvement in food hygiene for the upliftment of health standard of individuals. Engaging Healthcare workers, Veterinary extension officers and environmental health workers in a “One-health” approach should be encouraged to monitor the progress in identifying and preventing microbial contamination of the products. Health workers and the general public need to be enlighten on the zoonotic potentials of this organism and importance of strict hygiene practices in controlling its transmission.
The authors have not declared any conflict of interests.
REFERENCES
|
Akinyemi ERKO, Oyefolu AO, Opere B, Otunba-Payne VA, Oworu AO (1998). Escherichia coli in patients with acute gastroenteritis in Lagos, Nigeria. East Africa Medical Journal 75:512-515.
|
|
|
|
Al-Humam NA (2019). Detection of Escherichia coli, Salmonella spp. and Staphylococcus aureus in Ready-to-Eat Food in Al-Ahsa Province. Saudi Arabian Journal of Nutrition and Food Sciences 9:754.
|
|
|
|
|
Al-Zogibi OG, Mohamed MI, Hessain AM, El-Jakee JK, Kabli SA (2015). Molecular and serotyping characterization of shiga-toxogenic Escherichia coli associated with food collected from Saudi Arabia. Saudi Journal of Biological Sciences 22(4):438-442.
Crossref
|
|
|
|
|
Atikson RM, Besser JM, Bopp CA, Carlson C, Crandall C, George K, Gerner-Smidt P, Gladbach S, Gould LH, Hartley C, Maguire H (2012). Guidance for Public Health Laboratories on the Isolation and Characterization of Shigatoxin-producing Escherichia coli (STEC) from Clinical Specimens. Centers for Disease Control and Prevention and the Association of Public Health Laboratories.
|
|
|
|
|
Castro VS, Carvalho RCT, Conte-Junior CA, Figueiredo EES (2017). Shiga-toxin producing Escherichia coli: pathogenicity, supershedding, diagnostic methods, occurrence, and foodborne outbreaks. Comprehensive Reviews in Food Science and Food Safety 16(6):1269-1280.
Crossref
|
|
|
|
|
Chahed A, China B, Mainil J, Daube G (2006). Prevalence of Enterohemorrhagic Escherichia coli from serotype 0157 and other attaching and effacing Escherichia coli on Bovine carcasses in Algeria. Journal of Applied Microbiology 101(2):361-368.
Crossref
|
|
|
|
|
Chapman PA, Cerdan-Malo MA, Ellin M, Ashton R, Harkin MA (2001). Escherichia coli O157 in cattle and sheep at slaughter, on beef and lamb carcasses and in raw beef and lamb products in South Yorkshire, UK. International Journal of Food Microbiology 64:139-150.
Crossref
|
|
|
|
|
Effer EM, Isaacson L, Arntzen R, Heenan P, Canter T, Barette L, lee C, Mambo W, Levine A, Zaidi PMG (2001). Factors contributing to the emergence of Escherichia coli O157 in Africa. Emerging Infectious Diseases 7:812-819.
Crossref
|
|
|
|
|
Hussein HS, Bollinger LM (2005). Prevalence of Shiga-toxin producing Escherichia coli in beef cattle. Journal of Food Protection 68:2224-2241.
Crossref
|
|
|
|
|
Interagency Food Safety Analytics Collaboration (IFSAC) (2019). Food-borne illness source attribution estimates for 2017 for Salmonella, Escherichia coli O157, Listeria monocytogenes, and Campylobacter using multi-year outbreak surveillance data, United States. GA and D.C.: U.S. Department of Health and Human Services, CDC, FDA, USDA-FSIS.
|
|
|
|
|
Junior JC, Tamanini R, Soares BF, de Oliveira AM, de Godoi Silva F, da Silva FF, Augusto NA, Beloti V (2016). Efficiency of boiling and four other methods for genomic DNA extraction of deteriorating spore-forming bacteria from milk. Semina: Ciˆencias Agr'arias 37(5):3069-3078.
Crossref
|
|
|
|
|
Ministry of Animal Health and Fisheries (MAHF) (2012). Ministry of Animal Health and Fisheries, Sokoto State, Nigeria. Government Printer Sokoto.
|
|
|
|
|
Mukhufhi NS, Ori P, Peta MFR, Marais SJF, Moagiemang M, Venter AJC, Kabongo Pand Michel AL (2004). VTEC O157 in Slaughter Animals in the Gauteng Province of South Africa (S-B05). Proceed. 5th World Congress of Foodborne Infections and Intoxications 2:483-488. Berlin, Germany.
|
|
|
|
|
National Bureau of Statistics, Nigeria (NBS) (2017). Demographic Statistics Bulletin. National Bureau of Statistics. Abuja-Nigeria.
|
|
|
|
|
Olorunshola ID, Smith ST, Coker AO (2000). prevalence of Enterohaemmorhagic Escherichia coli O157:H7 in patients with diarrhea in Lagos Nigeria. Annals of Pakistan Institute of Medical Sciences 108:761-763.
Crossref
|
|
|
|
|
Paton JC, Paton AW (1996). Pathogenesis and diagnosis of Shiga-toxin producing E. coli infection. Clinical Microbiology Review 11: 450-479.
Crossref
|
|
|
|
|
Reuben A, Treminio H, Arias ML, Villalobos L (2002). Isolation of Escherichia coli O157:H7 from Costa Rican food. Review in Biomedicine13:273-276.
Crossref
|
|
|
|
|
RIMS (1991). Report of National Livestock Survey. FDLPCS. Federal Ministry of Agriculture, Nigeria.
|
|
|
|
|
Safarikova M, Safarik I (2001). Immunomagnetic separation of E. coli 026, 0111 and 0157 from vegetables. Applied Microbiology 33:36-39.
Crossref
|
|
|
|
|
Schaeffer EM (2012). Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany 365(8):709-717.
|
|
|
|
|
Vuddhakul V, Patararungrong N, Pungrasamee P, Jitsurong S, Morigaki T, Asai N, Nischibushi M (2000). Isolation and characterization of Escherichia coli O157 from retail beef and bovine faeces in Thailand. FEMS Microbiol Letters 182(2):343-347.
Crossref
|
|
|
|
|
Xia X, Meng J, McDermott PF, Ayers S, Blickenstaff K, Tran TT, Abbott J, Zheng J, Zhao S (2010). Presence and characterization of Shiga toxin -producing Escherichia coli and other potentially Diarrheagenic E. coli strains in Retail Meats. Applied and Environmental Microbiology 76:1709-17.
Crossref
|
|
|
|
|
Yakubu Y, Shuaibu AB, Ibrahim AM, Hassan UL, Nwachukwu RJ (2018). Risk of Shiga Toxigenic Escherichia coli O157:H7 Infection from Raw and Fermented Milk in Sokoto Metropolis Nigeria. Journal of Pathogens Article ID 8938597, 5 pages.
Crossref
|
|