ABSTRACT
More than half of the world’s population, especially women and children in the developing countries suffer from micronutrient malnutrition especially deficiency in iron and zinc. Micronutrient malnutrition problems increased the interest of researchers to increase the mineral contents (Fe and Zn) in cereals to ensure adequate attainment of dietary minerals. A lot of variability does exist for micronutrients (Fe, Zn, Vitamin A, etc.) content and bioavailability in many crops including rice. The current study was conducted to assess the variability for iron and zinc content in dehusked rice grains to identify mineral-rich families. This study was conducted with the major objectives of analysis of genetic variability for quality traits for grain iron and zinc content. Based on mean, genotypic coefficient of variability (GCV) and phenotypic coefficient of variability (PCV), heritability and genetic advance, it was understood that the progenies of ADT 37 × IR68144-3B-2-2-3 would be more useful for improving grain iron content with the desirable quality traits viz., kernel length, kernel breadth after cooking,. Similarly TRY (R) 2 × Mapillaisamba segregants could be used for improving the grain zinc content and breadth wise expansion ratio.
Key words: Genotypic coefficient of variability (GCV), phenotypic coefficient of variability (PCV), genetic advance (GA), iron and zinc content.
Rice is the life and the prince among cereals as this unique grain helps to sustain two thirds of the world’s population. Asia is the biggest rice producer, accounting for 90% of the world’s production and consumption of rice (Anonymous, 2007). It is considered as the main staple food for more than 50% of the world’s population. Rice provides 75% of the calories and 55% of the protein in the average daily diet of the people although it has an incomplete amino acid profile and comprises confined amount of essential micronutrients (Bhuiyan et al., 2002). The per capita consumption of rice is very high ranging from 62 to 190 kg/year (Graham et al., 1999). To be healthy, human beings require more than 20 mineral elements and more than 40 nutrients, particularly vitamins and essential amino acids, all of which can be supplied by an appropriate diet (Philip and Martin, 2005).However, human diets often lack one or more of essential nutrients and cause micronutrient malnutrition. Micronutrient malnutrition is recognized as a massive and rapidly growing public health issue especially among poor people living on an unbalanced diet dominated by a single staple grain such as rice. Among the major micronutrient deficiencies common in rice consuming countries, iron and zinc deficiencies (the so called “hidden hunger”), affect over three billion people worldwide, mostly in developing countries (Welch and Graham, 2004). Plant breeding to enhance the nutrient quality of staple food crops holds promise for a low cost and sustainable approach to alleviate the problem of micronutrient malnutrition among the poorest segments of the population of developing countries. This approach has been called biofortification. Biofortification reduces malnutrition by breeding essential micronutrients into staple crops. This approach bridges the fields of human nutrition, crop science and public health to develop a set of highly sustainable nutrition interventions in a cost-effective manner. Exploiting the genetic variation in crop plants for micronutrient content is one of the most powerful tools to change the nutrient balance of a given diet on a large scale.
Seeds of F3 generation of four cross combinations generated from Anbil Dharmalingam, Agricultural College and Research Institute, Trichy. viz., ADT 37 x IR68144-3B-2-2-3, TRY (R) 2 x Mapillaisamba were utilized as the experimental material in the present study. Among the parents viz., TRY (R) 2 and ADT 37 are high yielding commercial varieties and IR68144-3B-2-2-3 is a iron donor and Mapillaisamba is a zinc donor which were used in earlier hybridization programme for introgression of high iron and zinc contributing genes. The experiment was conducted at Agricultural College and Research Institute, Madurai. The F4 generation was raised during August to November, 2011 and F5 generation during December 2011 to April 2012 respectively. The F4 progenies were raised along with their parents in randomized block design with two replications. A total of five families were selected from each cross combination based on high iron and zinc content in F3 population. For each family, 75 seedlings per replication were raised with a spacing of 20 cm between the rows and 15 cm between the plants. Each family had five rows of 15 single plants each. The recommended agronomic practices were followed throughout the crop growth period. Five single plants per family per replication were randomly selected and forwarded as single plant progeny row in F5 generation. The mean data after computing for each character subjected to standard method of analysis of variance following Panse and Sukhatme (1961), phenotypic and genotypic coefficient of variation, heritability (Broad sense) and genetic advance as percent of mean were estimated by the formula as suggested by Burton (1952) and Johnson et al. (1955). The following quality traits are:
1. Kernel Length (KL): Ten unbroken hulled kernels with their tips intact from each genotype were measured for their length using vernier calipers. Average of length was taken in millimeters.
2. Kernel breadth (KB): Ten unbroken hulled kernels with their tips intact from each genotype were measured for their breadth using vernier callipers. Average of breadth was taken in millimetres.
3. Kernel length / breadth ratio (KLBR): The ratio between kernel length and breadth was estimated.
4. Kernel length after cooking (KLAC): Ten randomly selected whole milled rice kernels were taken in a labelled test tube and soaked in water for ten minutes. The tubes were placed in a water bath maintained at boiling temperature (100°C) for 20 min. After cooking, the cooked kernels (intact on both ends) were measured using a graph paper mounted in a glass frame and the average kernel length after cooking was expressed in millimetres.
5. Kernel breadth after cooking (KBAC): Ten randomly selected whole milled rice kernels were taken in a labelled test tube and soaked in water for ten minutes. The tubes were placed in a water bath maintained at boiling temperature (100°C) for 20 min. After cooking, the cooked kernels (intact on both ends) were measured using a graph paper mounted in a glass frame and the average kernel breadth after cooking was expressed in millimetres.
6. Linear elongation ratio (LER): The ratio of mean length of cooked rice to mean length of milled rice was computed as linear elongation ratio (Juliano and Perez, 1984):
In the present investigation ADT37 x IR68144-3B-2-2-3 showed high mean value for the characters viz., kernel length, kernel breadth after cooking, iron content. TRY (R) 2 x Mapillaisamba had high mean value for the characters viz., zinc content, breadth wise expansion ratio. The genotypic and phenotypic coefficient of variability were low in both the crosses for the most of the traits viz., kernel length, kernel breadth, kernel L/B ratio, kernel length after cooking, kernel breadth after cooking, linear elongation ratio and breadth wise expansion ratio (Table 1). It indicated that all the crosses might have attained homozygosity in F4 itself for these traits. These results were in accordance with Nandeshwar et al. (2010) who studied F2 progenies which was derived from the crosses IR-62 x Samba mashuri and Kunti x Dudheswar and the report of Kumar et al. (2006) in rice F2 generation for kernel length. kernel breadth recorded low genotypic and phenotypic coefficient of variability in both the generations for all the crosses. Similar findings were already have been reported by Umadevi et al. (2010) in 110 rice genotypes.
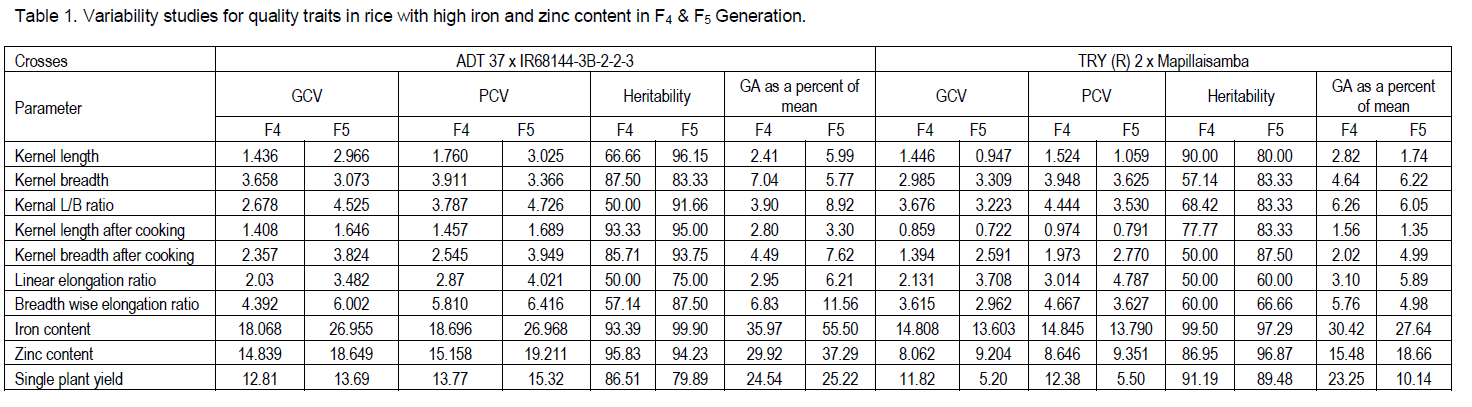
The findings of Vanaja and Babu (2006) recorded low genotypic and phenotypic coefficient of variability for kernel length after cooking and kernel breadth after cooking, breadth wise expansion ratio in 10 quality parameters in set of 56 high yielding diverse rice genotypes which is corroborates the current investigation. For iron and zinc content moderate GCV and PCV was observed in F5 generation. Hence, these traits need one or more generations in order to attain homozygosity. These results were in parallel with the findings of Kalaimaghal (2011) who observed moderate GCV and PCV for iron content in the in F2 and F3 the cross ADT37 x IR68144-3B-2-2-3.
High heritability coupled with high genetic advance was observed in ADT37 x IR68144-3B-2-2-3 only for iron and zinc content, except TRY(R)2 x Mapillaisamba which showed high heritability with moderate genetic advance. Hence, these traits were least influenced by environment and mostly governed by additive gene action. Therefore, there is scope for improvement of iron and zinc content by exercising selection pressure on these two characters. These results were in agreement with Shanmuga sundara pandian (2007), Purusothaman (2010) and Aswini Shamak et al. (2011) in F4 and F5 generation. Low heritability with low genetic advance as percentage of mean was observed for kernel length, kernel breadth and kernel L/B ratio in all the crosses in both the generations. The findings of Jayasree (2007) and Umadevi et al. (2010) in genotypes observed medium to high heritability with high genetic advance as percentage of mean for kernel length after cooking, kernel breadth after cooking, linear elongation ratio and breadth wise expansion ratio which in contrast with the result of current investigation. Based on mean, GCV and PCV, heritability and genetic advance, it was understood that the progenies of ADT 37 x IR68144-3B-2-2-3 would be more useful for improving grain iron content with the desirable quality traits viz., kernel length, kernel breadth after cooking. Similarly TRY (R) 2 x Mapillaisamba segregants could be used for improving the grain zinc content and breadth wise expansion ratio.
The authors have not declared any conflict of interest.
REFERENCES
Graham RD, Senadhira D, Beebe S, Iglesias C, Monsterio I (1999). Breeding for micronutrient density in edible portions of stable food crops: Conventional approaches. Field Crops Res. 60:57-80.
Crossref |
|
|
|
Jackson ML (1973). Soil and plant analysis. Prentice Hall of India Private Limited, New Delhi. |
|
|
Johnson HW, Robinson HF, Comstock RE (1955). Estimates of genetic and environmental variability in soybean. Agron. J. 47:314-318.
Crossref |
|
|
|
Jayasree (2007). Characterization of landraces, cultivars and new plant types of rice for quantitative and grain quality traits. M.Sc., (Ag.) Thesis (Unpubl.), TNAU, Coimbatore. |
|
|
|
Kalaimaghal R (2011). Studies on genetic variability of grain iron and zinc content in F2, F3 generation of rice (Oryza sativa L.). M.Sc. (Ag.) Thesis (Unpubl.), TNAU, Coimbatore. |
|
|
|
Kumar S, Gautam AS, Chandal C (2006). Estimates of genetic parameters for quality traits in rice (Oryza sativa L.) in mid hills of Himachal Pradesh. Crop Res. 32(2):206-208. |
|
|
|
Nandeshwar BC, Pal S, Senapati BK, De DK (2010). Genetic variability and character association among biometrical traits in F2 generation of some Rice crosses. Electron. J. Plant Breed. 1(4):758-763. |
|
|
|
Panse VG, Sukhatme PV (1961). Statistical methods for agricultural workers. 2nd edition, Indian Council. Agric. Res. New Delhi. P. 22. |
|
|
Philip JW, Martin RB (2005). Biofortifying crops with essential mineral elements. Trends Plant Sci. 10:586–593.
Crossref |
|
|
|
Purusothaman R (2010). Genetic analysis for high iron and zinc content in rice (Oryza sativa L.) grains. M.Sc. (Ag.) Thesis (Unpubl.), TNAU, Coimbatore. |
|
|
|
Shanmuga SG (2007). Studies on diversity and Genotype × Environment interaction for micronutrient content in rice (Oryza sativa L.). M.Sc. (Ag.) Thesis. (Unpubl.), TNAU, Coimbatore. |
|
|
|
Umadevi M, Veerabadhiran P, Manonmani S, Shanmugasundaram P (2010). Physico-chemical and cooking characteristics of rice genotypes. Electron. J. Plant Breed. 1(2):114-123. |
|
|
Welch RM, Graham RD (2004). Breeding for micronutrients in staple food crops from a human nutrition perspective. J. Exp. Bot. 55:353-364.
Crossref |