ABSTRACT
Barley nurseries comprising 820 lines with 479 unique pedigrees sourced from the International Centre for Agricultural Research in the Dry Areas (ICARDA) were screened for seedling and adult plant resistance (APR) against Australian isolates of barley leaf rust pathogen Puccinia hordei Otth. Ninety three percent of the lines were postulated to carry the seedling leaf rust resistance gene Rph3 based on their susceptibility in the greenhouse and field against Rph3 virulent pathotype and resistance to Rph3 avirulent pathotypes. The remaining lines showed either presence of uncharacterised seedling resistance (1%) and uncharacterised APR (1%). Five percent of lines were susceptible at both seedling and adult plant growth stages. Of the six lines identified to carry uncharacterised APR, three likely carried Rph20 based on the presence of the Rph20-linked marker bPb-0837. The results suggested that most of the ICARDA germplasm tested is not suitable for leaf rust resistance in Australia due to the presence of virulence for Rph3. Lines carrying uncharacterised seedling resistance and APR are potentially new sources of resistance, and are recommended for genetic analysis.
Key words: Hordeum vulgare, Puccinia hordei, adult plant resistance, Rph3.
Cultivated barley (Hordeum vulgare L. subsp. vulgare) is an important cereal crop (Ullrich, 2011) which ranks fourth in the world’s production after wheat, maize and rice (Schulte et al., 2009).
It is grown widely in Australia, where it is an important multi-billion dollar industry. The gross value of barley production in Australia is, however, hampered by many constraints, of which diseases alone account for an estimated average annual loss of $252 million (Murray and Brennan, 2010). Of the diseases that afflict barley, leaf rust (caused by Puccinia hordei Otth.) is considered to be most destructive in many parts of the world (Clifford, 1985). Significant losses due to leaf rust epidemics have been reported in Australia, New Zealand, Europe and USA (Murray and Brennan, 2010; Arnst et al., 1979; Cotterill et al., 1992; Griffey et al., 1994; Melville et al., 1976). Many of the known seedling leaf rust resistance genes have been rendered ineffective by the emergence of new pathotypes (pts) of P. hordei with matching virulence (Park, 2003).
During the years 1992 to 2001, eight new pts, each virulent for Rph12, were detected in Australia (Park, 2008) and recently, following the release of several barley cultivars with Rph3, a new pathotype (pt) with
virulence matching Rph3 (5457P+) was detected (Park, 2010). Currently, only six seedling resistance genes Rph7, Rph11, Rph14, Rph15, Rph18 and lately mapped Rph21 (Sandhu et al., 2012) are effective in Australia. In this context, several previous studies (Golegaonkar et al., 2009; Park, 2003, 2008) stressed the need to identify new sources of resistance to leaf rust in barley, including APR. Seedling resistance gene, temporally designated RphMBR1012, conferring resistance to the most virulent European leaf rust pathotypes, was mapped to the telomeric region of chromosome 1HS (König et al., 2012).
The first gene conferring APR to leaf rust in barley, Rph20, was mapped on chromosome 5HS (Hickey et al., 2011). Two markers linked to Rph20, EBmag0833 and bPb-0837, were reported by Liu et al. (2010), who proposed the use of bPb-0837 in marker assisted selection for APR against P. hordei. Recently, Singh et al. (2015) mapped the second APR gene Rph23 on chromosome 7H in H. vulgare which provides additive resistance against P. hordei under field conditions.
In addition to barley leaf rust, in the presence of heavy inoculum, stem rust caused by either Puccinia graminis Pers. f. sp. tritici Eriks., E. Henn., P. graminis Pers. f. sp. secalis Eriks., E. Henn., or the scabrum rust (Park, 2008), can affect barley in Australia. The barley stripe rust pathogen Puccinia striiformis f. sp. hordei does not occur in Australia (McIntosh et al., 2001; Park, 2008) but P. striiformis f. sp. pseudo-hordei (barley grass stripe rust; BGYR, Wellings, 2011; Wellings et al., 2000), can also infect some barley genotypes and wild barley grass in Australia.
The rust resistance of entries in several recent International Barley Observation Nurseries (IBONs) developed at ICARDA were examined in an attempt to identify potentially new sources of seedling resistance and APR to leaf rust. Four different IBONs (31st to 34th), released from 2003 to 2006, were examined.
Plant materials
Eight hundred and twenty lines representing four IBONs were introduced from ICARDA (Table 1), and screened for rust response in the greenhouse and field. The original nurseries were provided by the international nurseries program at ICARDA, Aleppo, Syria. Set of differential lines carrying seedling genes was used as controls for P. hordei (Table 4) as described by Park (2003) and Sandhu et al. (2012).
Pathogen material
Different pts of P. hordei were sourced from the rust collection maintained in liquid nitrogen at the Plant Breeding Institute (PBI), University of Sydney. For seedling tests, two pts of P. hordei (5457P+ and 5652P+) were used. In field testing, the predominant P. hordei pts were 5652P+ (2007 and 2008) and 5457P+ (2009). For multipathotype tests, four additional pts (200P-, 253P-, 5610P+ and 5653P+ +Rph13) were used. The virulence of the pts against seedling resistance (Rph) genes is detailed in Table 2.
Greenhouse screening
For greenhouse tests, all lines along with differential sets were planted in pots filled with the mixture of fine bark and coarse sand and fertilized using “Aquasol®” (100 g per 10 L of water per 200 pots) prior to sowing. Seedlings of differentials and barley lines were raised in 9 cm diameter pots by sowing four clumps (test lines) or five clumps (differentials) of each genotype using 8 to10 seeds per clump. Following sowing, pots were kept in a growth room at 20±2°C for germination. Seven-day old seedlings were fertilised with granular urea using “Incitec Pivot” w/w 46% nitrogen (50 g per 10 L of water per 200 pots). Seedlings at the one and a half leaf growth stage (9 to 10 days old) were inoculated and incubated according to the methods described by Sandhu et al. (2012). The seedlings were then transferred to naturally well lit microclimate rooms maintained at 23±2°C and scored 10 to 12 days after inoculation using 0 to 4 scale as described by Park and Karakousis (2002).
Field screening
All lines were tested at the field site Karalee. The lines were hand-sown as hill plots (30 to 40 seeds/plot) using 30 cm spacing during mid to late June in 2007, 2008 and 2009. A row of the susceptible cultivar Gus was sown as a rust spreader after every five hill plots of barley lines to allow the build-up and uniform distribution of inoculum. Four weeks after sowing, plots were fertilised using granular urea “Incitec Pivot” w/w 46% nitrogen @ 100 kg/hectare followed by irrigation. Plots were irrigated once a week or as required, using fixed sprinklers.
Field epidemics of leaf rust were created following the procedures described by McIntosh et al. (1995). Urediniospores (30–40 mg) were suspended in 1.5 L of light mineral oil (Shellsol®, Mobil Oil) and sprayed over buffer/spreader lines with an ultra-low-volume applicator (Microfit®, Micron Sprayer Ltd., UK). Four to five inoculations were performed during late evening on days that had a strong forecast of overnight dew. On the first and second inoculations, hot spots of disease were established by watering and covering small areas of the rust spreader with plastic hoods overnight to ensure adequate dew formation in case natural dew formation did not occur. Leaf rust was scored at the flag leaf growth stage in all three seasons using a modified Cobb’s scale (Peterson et al., 1948). Percentage of leaf area infected was followed by different scales; Immune (0), Resistant (R), Resistant to Moderately Resistant (R–MR), Moderately Resistant (MR), Moderately Susceptible (MS) and Susceptible (S).
Molecular analysis
Genomic DNA was extracted from leaf tissue of seedlings of barley lines characterised with APR as per techniques described by Bansal et al. (2010). The molecular marker bPb-0837 (Liu et al. 2010), closely linked to the APR leaf rust resistance gene Rph20 was used to genotype the lines selected from different nurseries. Ten micro litres of PCR reaction contained 2.0 µl of genomic DNA (50 ng), 1.0 µl of dNTPs (0.2 mM), 1.0 µl of 10x PCR buffer (Immobuffer, including 15 mM MgCl2), 0.25 µl of each forward and reverse primer (10 µM), 0.04 µl of Taq DNA (500 U Immolase DNA polymerase from Bioline) and 5.46 µl of ddH2O. The PCR amplification profile comprised an initial denaturation step at 95°C for 10 min, followed by 35 cycles of 30 s denaturation at 94°C, 60 s annealing at 55°C, 60 s extension at 72°C and a final extension step of 5 min at 72°C. Reaction was performed in a 96-well DNA theromocycler (Eppendorf Mastercycler, Germany).
PCR products were mixed with 3.0 µl of formamide loading buffer (98% formamide, 10 mM EDTA (pH 8.0), 0.05% (wt/vol) Bromophenol blue and 0.05% xylene cyanol). Two percent agarose gels were prepared by adding 2.0 gm agarose (Bioline) per 100 ml of 1x Tris-borate EDTA (TBE) buffer (90 mM Tris-borate + 2 mM EDTA-pH 8.0). For staining, 1.0 µl of ethidium bromide was added per 100 ml of gel solution. The gel solution was poured into moulds and allowed to cool for 40 min at room temperature. Eight to 10.0 µl of PCR product including loading buffer was loaded per well. One kb DNA marker HyperLadder™ IV (Bioline) was used as reference. Electrophoresis was carried out at 110 V for 1.5 h, and the separated bands were visualised under ultra violet light unit fitted with a GelDoc-IT UVP Camera.
89% (728 out of 820) of the lines tested from the four nurseries were resistant to pt 5652P+ in seedling greenhouse tests and in adult plant field tests during 2007 and 2008. All 728 lines were susceptible in the greenhouse and in the field when tested with the Rph3 virulent pt 5457P+ in 2009, indicating the very probable presence of seedling gene Rph3 only in all of these lines (Table 3).
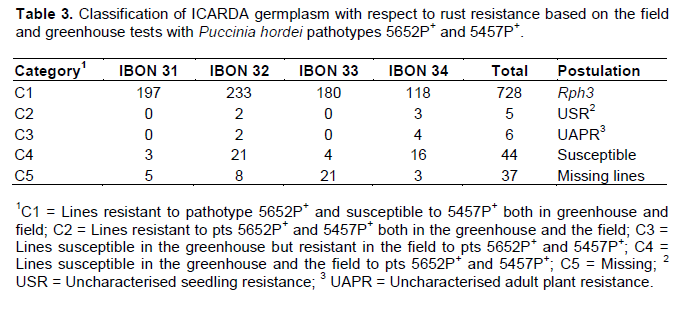
Five lines (IBON 32.34, IBON 32.126, IBON 34.88, IBON 34.95 and IBON 34.126) displayed seedling resistance in greenhouse tests to all pts, and were also resistant in the field against the two P. hordei pts used. Six lines (IBON 32.183, IBON 32.202, IBON 34.8, IBON 34.41, IBON 34.54 and IBON 34.110) were resistant only in the field (during all three years), indicating the presence of APR. Line numbers IBON 32.202, IBON 34.41, IBON 34.54 and IBON 34.110 showed resistant responses of R–MR to 10R at adult plant growth stages and line numbers IBON 32.183 and IBON 34.8 showed MR to MR–MS responses under field conditions (Table 5). Adult plant responses against P. hordei pt 5457P+ under field conditions are shown in Figure 3.
An overall analysis of the total 783 lines tested (excluding 37 missing lines) showed that 93% carried the major seedling resistance gene Rph3, 5.6% of the lines were susceptible at both growth stages, 0.65% of the lines were resistant to both P. hordei pts at both growth stages, and 0.75% of the lines possessed uncharacterised adult plant resistance (UAPR) as shown in Figure 1.
Multipathotype testing
The five lines in category C2 and six lines in category C3 (described in Table 3) were subjected to multipathotype tests in the greenhouse using six pts of P. hordei. Sets of differential lines included in these tests showed the expected infection types (ITs) against all six pts (Table 4).
Barley lines IBON 34.8, IBON 34.41, IBON 34.54, IBON 34.110, IBON 32.183 and IBON 32.202, which were resistant in the field, displayed high ITs against all six pts when tested in the greenhouse, indicating that they lacked detectable seedling resistance genes and confirming the presence of UAPR in all six lines. In contrast, lines IBON 32.34, IBON 32.126, IBON 34.88, IBON 34.95 and IBON 34.126, resistant in the field, also showed resistance in the greenhouse against all six P. hordei pts (Table 4).
Lines 88 and 95 from IBON 34 had the same pedigree and are hence sib-lines. Based on the resistance of these lines against the six pts used, they could carry either Rph5, Rph7, Rph11, Rph14, Rph15, Rph18, Rph21 or another uncharacterised seedling resistance (USR) gene. However, differences in the ITs produced by these lines in comparison with control genotypes carrying these genes suggested that the gene(s) present in each may be different. Differentials produced the expected ITs as shown in Figure 2.
Molecular marker analysis
Barley lines IBON 34.8, IBON 34.41, IBON 34.54, IBON 34.110, IBON 32.183 and IBON 32.202, which were resistant at adult plant stage in the field and displayed high ITs against all six pts at the seedling growth stage (Table 4), were selected for genotyping with the Rph20 linked marker bPb-0837 (Liu et al. 2010). Barley lines IBON 32.183, IBON 34.54 and IBON 34.110 amplified a 245 bp band, whereas no amplification occurred in lines IBON 32.202, IBON 34.8 and IBON 34.41 with marker bPb-0837. The control cultivars Pompadour, Baronesse, WI 3407 and Flagship amplified 245 bp bands, while no band was produced in tests with Stirling, Gus and Ricardo (Figure 4 and Table 5).
DISCUSSION AND CONCLUSION
Of the designated major seedling genes that confer resistance to P. hordei in barley (Rph1 to Rph21), only Rph7, Rph11, Rph14, Rph15, Rph18, Rph21 (Park, 2003; Park, 2010; Sandhu et al., 2012) and Rph20 (Hickey et al., 2011) are effective in Australia. It is well known that major genes can be easily overcome by new pts of P. hordei. This situation has occurred in Australia, with the frequency of virulence for Rph4 increasing following the widespread use of cultivar Grimmett carrying Rph4 (Cotterill et al., 1995), for Rph12 following the releases and widespread cultivation of barley cultivars including Franklin, Tallon, Lindwall and Fitzgerald, all of which carry Rph12 (Park, 2008) and more recently for Rph3, following the releases of cultivars Fitzroy, Yarra and Starmalt carrying Rph3 (Park, 2010). Plant breeders therefore have a limited choice in terms of resistance sources against P. hordei. In view of this, the present study sought new sources of resistance to P. hordei in four of the IBONs, which are distributed annually by ICARDA.
Tests of leaf rust response indicated that 44 lines were susceptible at both seedling and adult plant growth stages to all of the pts tested. During 2007 and 2008, more than 93% of the entries tested showed high levels of resistance to leaf rust in both seedling greenhouse and adult plant field tests. In early 2009, for the first time in Australia, virulence for the seedling resistance gene Rph3 was detected with the identification of a pt 5457P+ from the northern NSW (Park, 2010). When the nursery entries were tested with this new pt, 93% were susceptible in both the greenhouse and the field, strongly indicating that the resistance detected in previous tests was due to a single major gene, Rph3 and that no additional resistance was present in these lines. The occurrence of Rph3 only, in 93% of this germplasm indicated that it is highly vulnerable to leaf rust.
Five entries showed resistance to leaf rust that was effective in the field against pts 5652P+ and 5457P+ as
well as to a range of pts in the greenhouse (Table 4). Based on the ITs generated against a range of pts, it was postulated that these lines carry one or more unknown seedling resistance genes. While it is possible that these lines may carry one of the resistance genes (Rph5, Rph7, Rph11, Rph14, Rph15, Rph18 and or Rph21) that is effective against all of the pts used, this was considered unlikely because all five entries showed ITs that differed from those shown by all of the known effective genes except Rph18. It was considered unlikely that these nursery entries carried Rph18 because this resistance gene is derived from Hordeum bulbosum (Pickering et al., 2000) and has not yet been deployed in breeding programs. Genetic studies are therefore needed to characterise the seedling resistance identified in these five lines.
On the basis of multipathotype testing in the greenhouse and field testing for three consecutive seasons, six lines that carry APR to leaf rust were identified. It is known that many European cultivars carry APR to P. hordei (Golegaonkar et al., 2009; Park, 2008). Positive validation of the molecular marker bPb-0837 amplified 245 bp bands in lines IBON 32.183, IBON 34.54 and IBON 34.110 indicated the likely presence of the APR gene Rph20, reported on chromosome 5H (Hickey et al., 2011) and closely linked to this marker (Liu et al., 2010).
Similar amplification of 245 bp bands from DNA of reference stock; Pompadour, Baronesse, WI 3407 and Flagship by marker bPb-0837 (Liu et al., 2010) also supported the likely presence of Rph20 in genotypes IBON 32.183, IBON 34.54and IBON 34.110. The marker bPb-0837 failed to amplify a product in lines IBON 32.202, IBON 34.8 and IBON 34.41, indicating the likely presence of uncharacterised APR in each. These lines were resistant in the field and susceptible in the greenhouse to a range of P. hordei pts. The lines IBON 32.202 and IBON 34.41 showed identical field responses and it is possible that they might have a gene in common. The cultivar Ricardo was resistant in the field but failed to produce a PCR product when genotyped using marker bPb-0837, indicating the likely presence of an unknown resistance under field conditions. It will be useful therefore to undertake genetic analysis and allelic studies of unknown APR present in lines IBON 32.202, IBON 34.8 and IBON 34.41, along with Ricardo, to determine the mode of inheritance and their genetic relationship with the only other named APR gene for leaf rust in barley, Rph20 on chromosome 5H.
Given that virulence for seedling resistance gene Rph3 is now present in eastern Australia, 93% of the germplasm tested here carries Rph3 only is of limited value for leaf rust resistance. The diversity of leaf rust resistance among these four ICARDA nurseries is very narrow as only 11 lines (6 with unknown APR and 5 with unknown seedling resistance) were identified in the study. Out of six lines with APR, three likely carry Rph20. Eight lines (three with APR and five with seedling resistance) were identified that carry potentially uncharacterised resistance to leaf rust and are therefore potentially valuable as new sources of resistance.
It is recommended to undertake genetic analysis of these eight lines to understand their inheritance and genetic relationship with other known genes for their effective utilisation in breeding programs. The present studies again stress the importance of identifying new sources of leaf rust resistance to diversify the genetic base of resistance and for additional and better choice of resistance in breeding programs. At the same time, deployment of single major known effective genes (Rph7, Rph11, Rph14, Rph15, Rph18 and Rph21) should be conducted wisely by avoiding the release of cultivars with single major genes only. Efforts should be made to pyramid the resistance genes available to reduce the chance of matching virulence developing in pathogens. Given that only one gene conferring APR to leaf rust in barley has been characterised to date, the identification of one or more potentially new sources of APR could provide a means of achieving durable resistance via APR genes pyramiding.
The authors have not declared any conflict of interests.
The first author would like to sincerely thank and acknowledge the Australian Grains Research and Development Corporation for the provision of a Postgraduate Research Scholarship, and the support provided by the University of Sydney that enabled these studies to be initiated and completed. Material provided by ICARDA is greatly acknowledged.
REFERENCES
|
Arnst BJ, Martens JW, Wright GM, Burnett PA, Sanderson FR (1979). Incidence, importance and virulence of Puccinia hordei on barley in New Zealand. Ann. Appl. Biol. 92:185-190.
Crossref
|
|
|
|
Clifford BC (1985). Barley leaf rust. In: Roelfs AP, Bushnell WR (eds) The cereal rusts. Academic, Orlando. pp. 173-205.
Crossref
|
|
|
|
|
Cotterill PJ, Park RF, Rees RG (1995). Pathogenic Specialization of Puccinia hordei Otth. in Australia, 1966-1990. Aust. J. Agric. Res. 46:127-134.
Crossref
|
|
|
|
|
Cotterill PJ, Rees RG, Platz GJ, Dill-Macky R (1992). Effects of leaf rust on selected Australian barleys. Aust. J. Exp. Agric. 32:747-751.
Crossref
|
|
|
|
|
Golegaonkar PG, Singh D, Park RF (2009). Evaluation of seedling and adult plant resistance to Puccinia hordei in barley. Euphytica 166:183-197.
Crossref
|
|
|
|
|
Griffey CA, Das MK, Baldwin RE, Waldenmaier CM (1994). Yield losses in winter barley resulting from a new race of Puccinia hordei in North America. Plant Dis. 78:256-260.
Crossref
|
|
|
|
|
Hickey LH, Lawson W, Platz GJ, Dieters M, Arief VN, Germán S, Fletcher S, Park RF, Singh D, Pereyra S, Franckowiak J (2011). Mapping Rph20: A gene conferring adult plant resistance to Puccinia hordei in barley. Theor. Appl. Genet. 123:55-68.
Crossref
|
|
|
|
|
König J, Kopahnke D, Steffenson BJ, Przulj N, Romeis T, Röder MS, Ordon F and Perovic D (2012). Genetic mapping of a leaf rust resistance gene in the former Yugoslavian barley landrace MBR1012. Mol. Breed. 30(3):1253-1264.
Crossref
|
|
|
|
|
Liu F, Gupta S, Zhang X, Jones M, Loughman R, Lance R, Li C (2010). PCR markers for selection of adult plant leaf rust resistance in barley (Hordeum vulgare L.). Mol. Breed. 28:657-666.
Crossref
|
|
|
|
|
McIntosh RA, Bariana HS, Park RF, Wellings CR (2001). Aspects of wheat rust research in Australia. Euphytica 119:115-120.
Crossref
|
|
|
|
|
McIntosh RA, Wellings CR, Park RF (1995). Wheat Rusts: An Atlas of Resistance Genes. CSIRO, Canberra, Australia.
Crossref
|
|
|
|
|
Melville SC, Griffin GW, Jemmett JL (1976). Effects of fungicide spraying on brown rust and yield in spring barley. Plant Pathol. 25:99-107.
Crossref
|
|
|
|
|
Murray GM, Brennan JP (2010). Estimating disease losses to the Australian barley industry. Aust. Plant. Pathol. 39:85-96.
Crossref
|
|
|
|
|
Park RF (2003). Pathogenic specialization and pathotype distribution of Puccinia hordei in Australia, 1992 to 2001. Plant Dis. 87:1311-1316.
Crossref
|
|
|
|
|
Park RF (2008). Breeding cereals for rust resistance in Australia. Plant Pathol. 57:591-602.
Crossref
|
|
|
|
|
Park RF (2010). Annual report: Cereal rust survey 2009–2010, The University of Sydney. pp. 1-12.
|
|
|
|
|
Pickering RA, Malyshev S, Kunzel G, Johnston PA, Korzun V, Menke M, Schubert I (2000). Locating introgressions of Hordeum bulbosum chromatin within the H. vulgare genome. Theor. Appl. Genet. 100:27-31.
Crossref
|
|
|
|
|
Sandhu KS, Forrest KL, Kong S, Bansal UK, Singh D, Hayden MJ, Park RF (2012). Inheritance and molecular mapping of a gene conferring seedling resistance against Puccinia hordei in the barley cultivar Ricardo. Theor. Appl. Genet. 125:1403-1411.
Crossref
|
|
|
|
|
Schulte D, Close TJ, Graner A, Langridge P, Matsumoto T, Muehlbauer G, Sato K, Schulman AH, Waugh R, Wise RP, Stein N (2009). Update on the International Barley Sequencing Consortium, The International Barley Sequencing Consortium-At the Threshold of Efficient Access to the Barley Genome. Plant Pathol. 149:142-147.
|
|
|
|
|
Singh D, Dracatos P, Derevnina L, Zhou MX, Park RF (2015). Rph23: A new designated additive adult plant resistance gene to leaf rust in barley on chromosome 7H. Plant Breed. 134:62-69.
Crossref
|
|
|
|
|
Ullrich SE (2011). Barley: Production, improvement and uses. Blackwell, Oxford, UK.
|
|
|
|
|
Wellings CR (2011). Global status of stripe rust: a review of historical and current threats. Euphytica 179:129-141.
Crossref
|
|
|
|
|
Wellings CR, Burdon JJ, Mcintosh RA, Wallwork H, Raman H, Murray GM (2000). A new variant of Puccinia striiformis causing stripe rust on barley and wild Hordeum species in Australia. Plant Pathol. 49:803.
Crossref
|
|