ABSTRACT
Cassava brown streak disease (CBSD) is currently the major disease affecting cassava production in Eastern and Southern Africa. Breeding for resistance has been hampered by a lack of sources of resistance and the complexity of CBSD. This study was initiated to assess the possibility of exploiting inbreeding, as a strategy for generating new sources of resistance to CBSD. This was based on the premise that inbreeding increases the additive variance upon which selection for desirable phenotypes can be made. Eight cassava progenitors (S0): Namikonga, 182/006661, Kigoma Red, Tz/130, Tz/140, 130040, 0040 and 100142 were selfed for one generation to produce the first inbred generation (S1). The S1 progenies generated were evaluated for two seasons (seedling and clonal evaluation trial) in a high CBSD pressure area. Promising clones were re-evaluated to confirm their CBSD reaction status. Results obtained showed that within each family, a few S1 inbreds (1-15) had higher levels of resistance compared to the S0 progenitors with the highest number observed in Tz/130. It is possible therefore to get transgressive progenies through inbreeding.
Key words: Cassava brown streak disease, inbreeding, cassava partial inbreds, new sources of resistance, inbreeding depression, resistance breeding.
Cassava (Manihot esculenta Crantz.) is one of the most important root crops grown widely in tropical countries notably in sub-Saharan Africa, South America and Asia. In recent years, cassava production has been greatly hindered by a myriad of biotic stresses. Of these, cassava brown streak disease (CBSD) is the major disease affecting cassava production in Eastern and Southern Africa (Pennisi, 2010). The disease is caused by two virus species, cassava brown streak virus (CBSV) and Uganda cassava brown streak virus (UCBSV), both are Ipomoviruses of family Potyviridae characterized by an elongate flexuous filament 650 to 690 nm long (Monger et al., 2001; Mbanzibwa et al., 2011). The presence of two distinct species of virus that causes CBSD and a lack of natural resistance has posed a great challenge to breeding efforts tailored towards increasing cassava productivity in CBSD affected areas. These viruses are distributed in Tanzania (Ndunguru et al., 2015), Kenya (Munga, 2008), Uganda (Alicai et al., 2007), Democratic Republic of Congo (Mulimbi et al., 2012), Rwanda (Tomlinson et al., 2013), Burundi (Bigirimana et al., 2011), Malawi (Mbewe et al., 2015) and Mozambique (Zacarias and Labuschagne, 2010). Undocumented reports of CBSD outbreaks in Zambia have also been made.
Breeding for CBSD resistance is the most efficient way to combat the disease. The pioneering breeding program for cassava mosaic disease (CMD) and CBSD was started more than 70 years ago at Amani Research station in Tanzania. The program initially focused on searching for sources of resistance among different cassava genotypes. According to Jennings (1957), limited progress was made which led to the use of cassava wild relatives in the program. Several crosses were made between M. esculenta and wild Manihot species (Manihot glaziovii, Manihot melanobasis, Manihot cathartica, Manihot dichotoma and Manihot saxicola) in order to introgress CMD and CBSD resistance genes into preferred cassava genotypes (Hillocks and Jennings, 2003). Through interspecific hybridization and backcrossing several hybrids with reasonable levels of CBSD resistance, such as Namikonga (also known as Kaleso in Kenya) were developed and incorporated in the farming system. IITA have actively been breeding for CBSD resistance in Tanzania since 2004, incorporating germplasm derived from the Amani program.
Diallel studies of the inheritance of CBSD resistance/tolerance conducted in Kenya (Munga, 2008), Uganda (Tumuhimbise et al., 2014) and Tanzania (Kulembeka et al., 2012) have demonstrated the relative importance of additive genetic effects as opposed to non-additive effects). Zacarias and Labuschagne (2010) showed the importance of non-additive genetic effects in germplasm from Mozambique. Additivity presents the possibility for enhancing levels of resistance through the inbreeding of tolerant genotypes. According to Walsh (2005), inbreeding allows “concentration” of desirable genes originally present in the elite clone. By forcing an average of half of the loci to become homozygous, the additive value of a selfed individual or progeny is increased, and through selection, any resultant homozygous deleterious alleles can be purged (Barrett and Charlesworth, 1991). Inbreeding results in progeny at both fitness extremes, that is, extremely high fitness with many homozygous advantageous alleles with few deleterious mutations and extremely low fitness with many homozygous deleterious mutations. Indeed, a recent study on the segregation of selected agronomic traits in cassava inbreds (Kawuki et al., 2011) showed an increase in performance in agronomic traits (harvest index and root dry matter content) in some inbreds compared to their respective non-inbred parents. Here, it was hypothesized that S1 partial inbreds will not only be better progenitors but will also possess higher levels of resistance to CBSD than their respective non inbred parents. This study was initiated to generate and evaluate cassava partial inbred for resistance and/or tolerance to CBSD in Uganda.
Generation of S1 families from S0 parents
Ten cassava progenitors (S0): Namikonga, 182/006661, Kigoma Red, Tz/130, Tz/140, 130040, 0040, Kiroba, Nachinyaya and I00142 from Tanzania were selected after CBSD tolerance had been confirmed using quantitative real-time PCR diagnostics (Kaweesi et al., 2014) and established in isolated plots at National Crops Resources Research Institute (NaCRRI), Central Uganda. With the exception of Tz/130 and Tz/140, which were selected in Uganda from open pollinated seeds introduced from Tanzania in 2005, all other progenitors were introduced from Tanzania as stem cuttings in 2009. Each parental line was represented by 20 plants which were established in two-row plots of 10 plants. At flowering, selfing was done by hand according to standard procedures to generate partial inbred lines (S1). Within a cassava field, it is possible to get mature pollen and mature female flowers of the same clone (from different branches or plants) and thus selfing is possible. After selfing, flowers were bagged for at least 2 to 3 days to avoid contamination, labeled appropriately and the number of flowers selfed and the number of selfed fruits per plant were recorded. Any open pollinated flowers were removed to avoid mixtures. After three months, the mature fruits were harvested and numbers of seeds recorded. The harvested S1 seeds were established in a nursery at NaCRRI after a two month period to break dormancy. After two months in the nursery, the S1 seedlings were transplanted in a well-prepared field for CBSD evaluation.
S1 seedling evaluation trial
Eight S1 families were evaluated. All seedlings belonging to a single family were established in the same block. Spreaders using a CBSD – susceptible variety (TME 204) were planted after every four rows of test genotypes to augment the CBSD pressure. This trial was planted during the first rains (March - June) of 2011. Data for CBSD were collected on individual seedlings at two-month intervals after the third month after planting (MAP). Cassava raised from seed usually produces a few storage roots (1-10) (Tumuhimbise et al., 2014) which also provide an opportunity for CBSD root necrosis evaluation. However, subsequent evaluations were done on cloned genotypes, thus, after nine months, each plant in the seedling evaluation trial (SET) was individually harvested and data were taken for foliage yield, root yield, CBSD root severity and CBSD root incidence. Thereafter, 8 to 12 cuttings were taken from each parent (S0) and self (S1) to generate clones for further evaluation.
Evaluation of S1 clones for CBSD resistance
S1 clones were evaluated in clonal trials during 2012/13 and 2013/14. Clonal evaluation trials (CET) were established at NaCRRI using single rows of six plants per genotype. Both S1 progeny and the non-inbred parent (S0) were established in the CET. The first clonal trial was planted during the first rains (April) of 2012. Each row represented a single clone and the spacing was 1 m within and between the rows. To control variability in the field, clones from a given family were separated into three groups of roughly equal size and each group of a family was randomly allocated to one of the blocks along with respective parental genotypes for comparison. No selection was done; all seedlings were cloned and evaluated. Spreader rows of TME 204 were established between rows to augment CBSD pressure. The genotypes were grown for 12 months under rain fed conditions with no fertilizer or herbicide applied.
Above-ground CBSD symptoms (on leaves and stem) were assessed visually on every plant in each plot. Both incidence (proportion of cassava plants in a plot expressing CBSD symptoms) and severity (degree of infection of CBSD on the individual plant) were used to quantify the disease. Five data sets at three, five, seven, nine and eleven months after planting (MAP) were collected. A severity scale of 1 to 5 (Gondwe et al., 2003) was adopted for above ground symptoms where 1- no symptom, 2- mild symptom (1-10%), 3- pronounced foliar chlorotic mottle and mild stem lesion (11-25%), 4- severe chlorotic mottle and stem lesion (26-50%) and 5- very severe symptoms (>50%). Severity scores for root necrosis were also taken on all roots harvested per plot at 12 MAP. Severity scores for root necrosis were based on a 1-5 scale where 1- no necrosis, 2- mild necrotic lesions (1-10%), 3-pronounced necrotic lesions (11-25%), 4- severe necrotic lesions (26-50%) and 5- very severe necrotic lesion (>50%).
Clones that maintained a root severity of 1 or 2 were selected and re-evaluated at NaCRRI during the CET-2 established in 2013/2014 season to further confirm their resistance/tolerance levels. Thus, the S1 inbreds with scores of 1 or 2 were evaluated for three seasons.
Data analysis
Root severity scores were converted into disease severity mean (DSM) using the following formula:
Disease incidence (DI) of CBSD in harvested roots was quantified as a ratio of the number of roots showing roots symptoms to the total number of roots harvested per plant per genotype. Disease index of every clone was derived as a product of DI and DSM.
Data on disease index was subjected to one-way analysis of variance using Genstat (ver. 14) at a significance level of 5% to compare families. The field reaction of each generated partial inbred to CBSD was compared to that of the respective progenitor (S0) by subjecting disease index data for the family (S0 progenitor and its S1 inbreds) to the analysis of variance using Genstat (ver. 14) (Payne et al., 2011). To determine the effect of inbreeding, the disease index of each partial inbred was compared to the disease index of their respective parent to determine the total number of positively transgressive progenies per family. Each S1 partial inbred that had a lower disease index compared to its respective progenitor was considered a positively transgressive progeny. The percentage of positively transgressive progenies per family was compared to determine the best progenitors.
To measure the heritability of resistance or tolerance to CBSD, a parent-offspring regression was made using mean values of disease index of parents and offspring based on root necrosis data collected in one environment, NaCRRI. The offspring were regressed on that of their parent using standard linear regression model y1 = b0+b1x1 +e, where y1 is the mean of offspring of the ith family, b0 is the intercept, b1 is the regression coefficient and x1 is the parent of the ith family and e is the random error. The expression h2=2b1 was used since partial inbred families are regressed on a single parent. Parent-offspring regression analysis was performed using Genstat (ver. 14).
S1 seed germination and survival
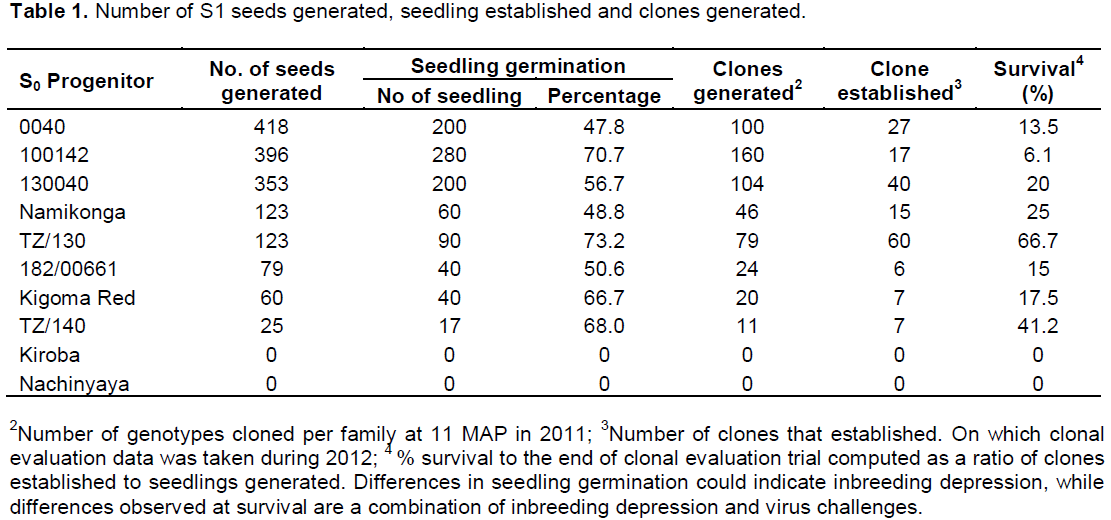
Ten cassava genotypes (S0) were evaluated and selfed to produce partial inbreds (S1). Of the ten SO progenitors, only eight were able to produce seeds in variable proportions while two genotypes (Kiroba and Nachinyaya) did not produce seed due to male sterility. Due to high heterozygosity of cassava as a crop, it had been hypothesized that a low rate of germination would be obtained due to inbreeding depression. Results show variable germination rates among the families, ranging from 47.8 to 73.2%. Under ideal conditions, the germination rate of non-inbred cassava population is expected to be 90 to 100%. Therefore, these results show moderate effects of inbreeding on germination.
A general reduction was observed in the survival rate in all the families between the SET and CET. Two S0 progenitors (0040 and 100142) that produced the highest number of seeds had the lowest survival rate at CET of 13.5 and 6.1%, respectively, as compared to TZ/130 and TZ/140 with a survival rate of 66.7 and 41.2% (Table 1). Though, this study intended to explore the benefits of inbreeding in search of resistance to CBSD, effects of inbreeding on fitness traits were noted. A large proportion of seedlings generated was characterized by a loss of vigour, height and reduction in growth, and therefore, did not survive to be advanced to the clonal evaluation trial. The low survival rate could be partly attributed to inbreeding depression and virus challenges (both CMD and CBSD). Inbreeding depression for sprouting, vigor, height, flowering, harvest index and dry matter content was low or absent in some families of the clones that were advanced to the CET (Supplementary Tables 1 and 2).
Response of partial inbreds to cassava brown streak disease
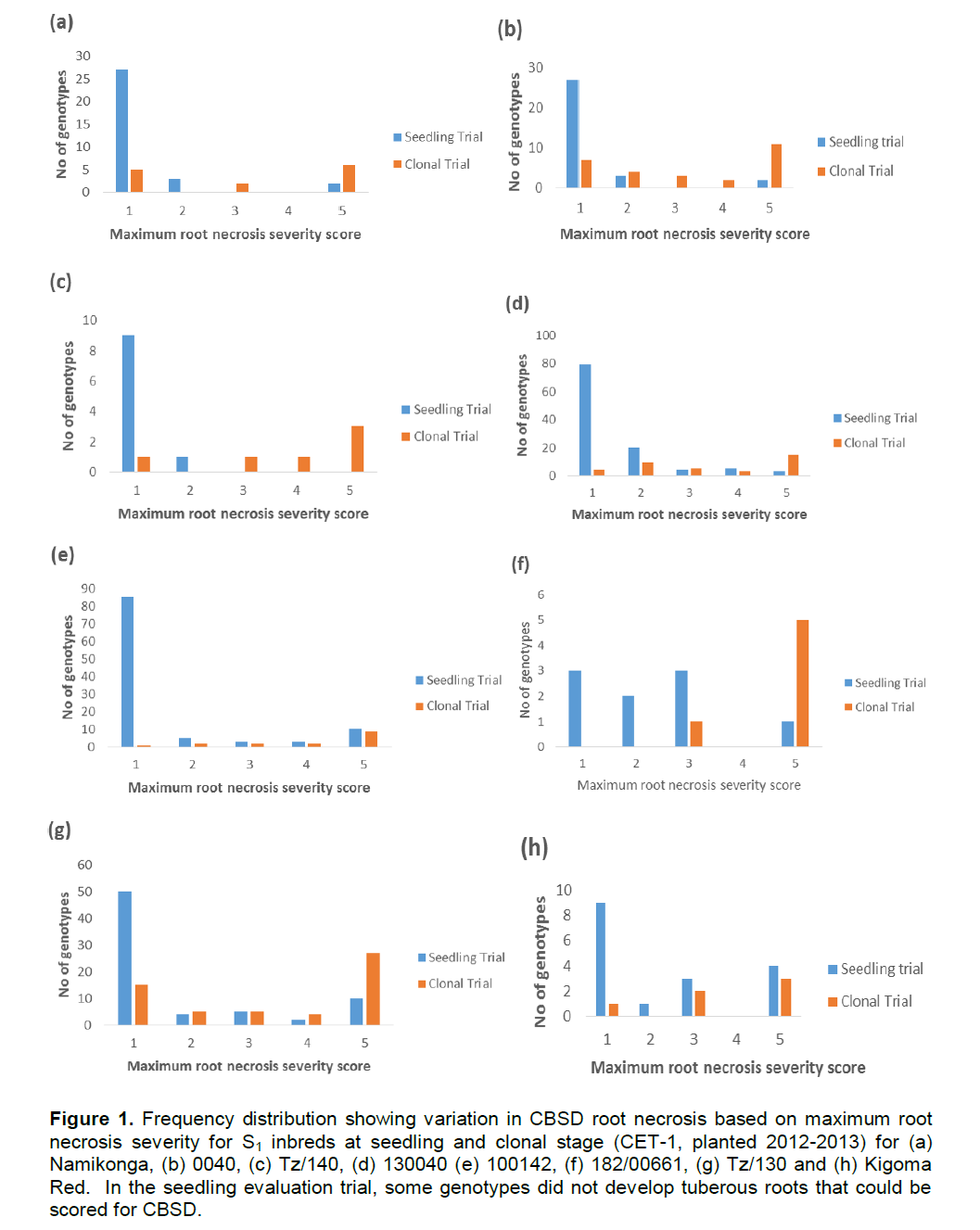
Both foliar and root symptoms were used to determine the response of the generated partial inbreds to CBSD. Variation in susceptibility to CBSD among different clones of different cassava families was striking. It ranged from 0 to 100% foliar incidence with a severity score of 1 to 4.5. All the parents showed foliar symptoms while a variable number of partial inbreds in each family remained symptomless (Table 2). A similar pattern was observed also for root symptom. Different phenotypic classes (partial inbreds with max root necrosis score 1, 2, 3, 4 and 5) for CBSD were observed differentially in the different families. At SET, most of the partial inbreds had a maximum score of 1 for root necrosis while family Kigoma Red and 182/00661 showed even distribution across classes (Figure 1). This distribution changed from the SET and CET-1 with increasing frequency of genotypes with maximum root severity scores 4 and 5 as compared to scores 1 and 2. On the other hand, inbreds from Namikonga exhibited two extremes, that is, 42% had score 1 (resistant), while 42% had score 5 (susceptible).
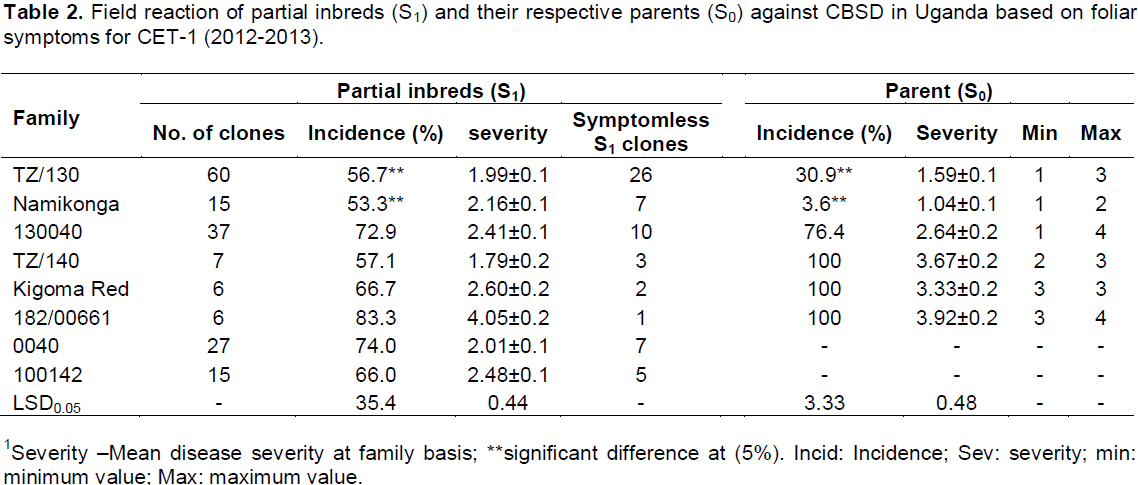
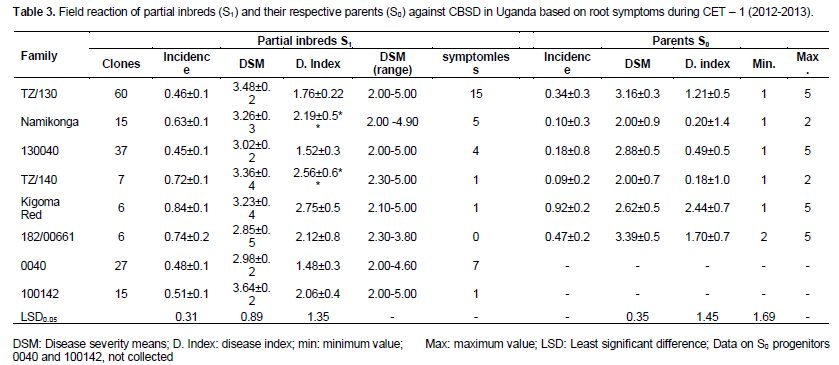
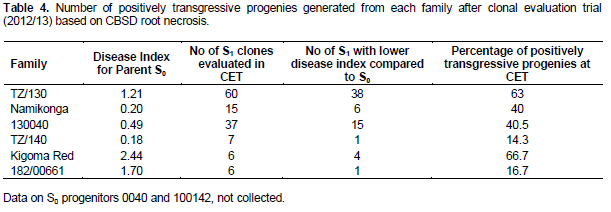
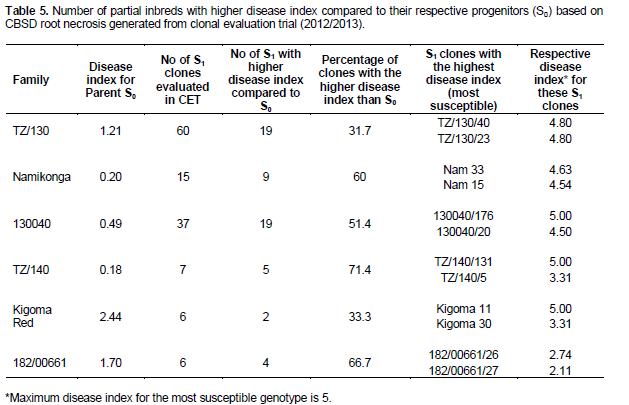
In comparison to S0 progenitors, there was no significant difference between the mean of all the generated partial inbreds in a given family compared to their respective S0 progenitors. However, there were partial inbreds that performed better than their respective parent based on both foliar and root symptoms as hypothesized. These were considered positively transgressive progenies. A small proportion (1-15) of partial inbreds generated from all families (except 182/00661) did not show both foliar and root symptoms after SET and CET. The highest percentage of partial inbreds that remained symptomless was obtained in family Namikonga and TZ/130 (Table 3). The TZ/130 and Kigoma Red families contributed a large percentage of positively transgressive progenies (Table 4). Contrastingly, some families like 182/00661 and TZ/140 did not perform as expected, with a higher number of inbreds with higher disease index than their respective progenitors. These parents produced higher percentages of negatively transgressive progenies (Table 5).
Re-evaluation of selected S1 partial inbreds in CET-2 (2013/2014)
When S1 partial inbreds with root severity, scores of 1 and 2 were re-evaluated in 2013/2014, some with a maximum score of 1 for root necrosis remained symptomless, while some maintained a very low incidence and maximum severity of 2. Of the 34 S1 partial inbreds that remained symptomless in 2012/2013, nine S1 inbreds remained symptomless for CBSD root necrosis, while six S1 inbred maintained a low incidence (1.25 to 7.96%) and a maximum severity of 2 (Table 6). The absence of root symptoms and/or limited symptom expression after three years of exposure to CBSD at Namulonge suggests the presence of elevated tolerance or resistance levels in S1 progenies compared to the parents which all showed some symptoms. These results show that inbreeding can not only enhance field resistance to CBSD, but can also be used as a strategy to generate new genetic stocks with high resistance level for CBSD resistance breeding. Offspring-parent regression analysis provided a linear model (y=0.216x + 1.752), with a slope of 0.216, thus providing an estimate of heritability across all the families of 0.43.
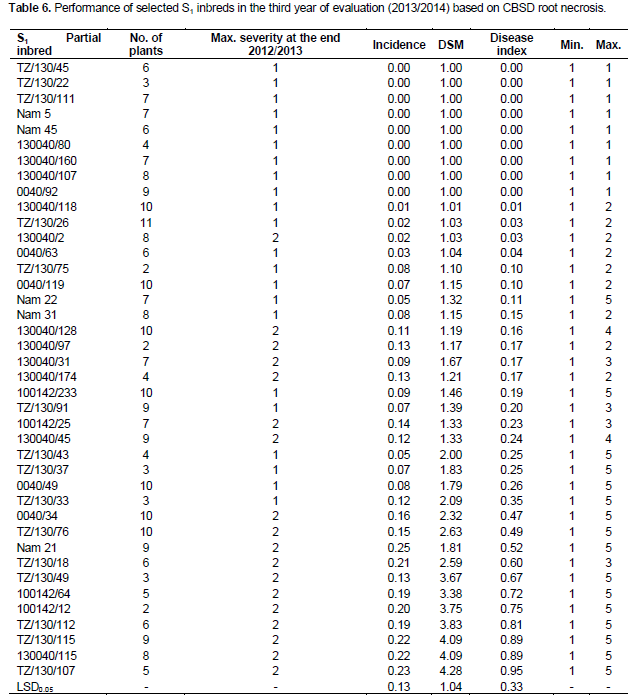
The objective of this study was to develop and screen S1 partial inbreds derived from some of the most tolerant genotypes to CBSV and UCBSV in Uganda. Most of these parental lines were sourced from Tanzania, where CBSD has been prevalent for over 50 years, and where an inter-specific breeding program for CMD and CBSD was conducted from the 1930s. Evaluations were done at the seedling and clonal stages in a CBSD hotspot at Namulonge, relative to their parents. Promising S1 partial inbred clones with a score of 1 and 2 were further evaluated in a replicated clonal trial for the third year (2013/2014).
A varying number of the generated S1 clones remained symptomless for both UCBSV and CBSV (on roots) for the two seasons evaluated in a “hotspot” zone (Tables 3 and 6). Within each family, a few S1 inbreds (1-15) showed higher levels of resistance than the S0 progenitors and are therefore considered positively transgressive progenies. These clones are potential sources of resistance to CBSD. Certainly, the absence of root symptoms and/or limited symptom expression after three years of exposure to CBSD at Namulonge suggests the presence of elevated tolerance or resistance levels in S1 progenies compared to the parents which all showed some symptoms.
Inbreeding increases homozygosity, thereby changing the distribution of genetic variation of a trait (in our case, resistance or tolerance to CBSD). This change increases the visibility of genetic variation to selection and also exposes the phenotypic effects of previously hidden recessives (both beneficial and deleterious) (Charlesworth, 1992). According to Kelly (1999a and 1999b), the extent of these effects depends on the pattern of dominance, linkage disequilibrium and allele frequency in the parent. Our study showed strong parental genotypic differences in inbreeding effects on CBSD resistance/tolerance which is likely to reflect the number of advantageous recessive CBSD resistance alleles in the heterozygous state in a parent, and dominance. For example, family TZ/130 had a comparatively higher percentage of positively transgressive progenies compared to other families. It is likely this parent possesses more loci influencing CBSD resistance in the heterozygous state. Therefore, selection of S0 progenitors is important for the success of an inbreeding program. Once outperforming S0 progenitors are identified based on partial inbred performance, a higher number of partial inbreds from the best performing S0 progenitors could be generated in addition to further selfing generations. This will increase the chances of generating more positive transgressive progenies with elevated levels of resistance to CBSD.
Previous studies conducted on the genetics of CBSD resistance (Munga, 2008; Kulembeka, 2010; Tumuhimbise et al., 2014) indicate that CBSD resistance is largely under the control of additive genetic effects. Additive effects can easily be exploited with inbreeding. From quantitative genetics theory, additive and non-additive genetic effects in S1 can be partitioned between and within families in the following proportions; between families (σ2A=1, σ2D=1/4) and within families (σ2A=1/2, σ2D=1/2). Consequently, total additive effects will be σ2A= 3/2, while total non-additive effects will be σ2D=3/4 (Hallauer and Miranda, 1981). This explains to some extent the higher levels of CBSD resistance observed in the few S1 clones. The positive transgressive segregation observed in some cassava families may also be due to the unmasking of advantageous recessive alleles that are heterozygous in parental lines and the additive action of these unmasked alleles. According to Kawuki et al. (2011), there was an increase in mean performance in amylose content among six cassava S1 families generated at NaCRRI. That study also observed S1 individuals with higher dry matter content and harvest index compared to S0. Desirable phenotypes have also been previously reported in S1 cassava (Ceballos et al., 2004, 2007). Inbreeding, therefore, presents opportunities to improve traits in cassava especially those that are quantitative in nature. Inbreeding has also been used in other crops to improve plant defense system against biotic stresses. One example is a study by Hall-Sanders and Eubanks (2005), in which inbreeding increased the resistance of Ipomea hederacea var intergniuscula to both specialist and opportunistic generalist herbivore among inbreds compared to the outcrossed.
The heritability estimate of 0.43 obtained in this study implies that only 43% of the observed phenotypic variance in response to CBSD among inbreds is due to additive genetic effects. This is a modest estimate which implies that the response of generated partial inbreds to CBSD can be predicted by severity or disease index of parental genotypes. Moderate estimates obtained in this study also suggest that substantial genetic gain would be obtained when selecting for resistance in partially inbred cassava families though selection would be more effective in later generations (S3 or S4).
The CBSD phenotypic class frequency distribution (Figure 1) observed in different families in this study showed that there was continuous variability in all families except for Namikonga which had two distinct classes (susceptible and resistant). Segregation implies that some resistance genes are in the heterozygous state; however, in Namikonga, it is likely that some dominance effects are also operational. Breeding will be easier if we know that resistance genes are fixed in the source genotype, however as soon as these are crossed, they would return to the heterozygous state, and would have to be backcrossed, selfed or intercrossed, to recover the homozygous state.
Therefore, if molecular markers were available that were associated with the quantitative trait loci (QTL) that confer the resistance, breeders could determine whether associated genes were in the homozygous or heterozygous states which would help in the accuracy of breeding. Quantitative trait loci (QTL) mapping experiments are underway to better track contributing alleles and their homozygous/heterozygous states during breeding.
Furthermore, there was variation in the symptom expression within a genotype in the SET and the CET (Figure 1). This variability could be due to virus multiplication and the accumulation over the first season, and carry over, through stakes to the second field season. This study shows the importance of screening cassava genotypes for more than one cycle in a hotspot to properly determine their response to CBSD, if starting with uninfected stakes or seedlings.
Low germination percentage could be attributed to inbreeding depression. In addition, low survival rates at the clonal stage could have resulted from a combination of inbreeding depression and cassava virus accumulation notably CMD. With selfing, some recessive deleterious alleles, once masked by dominance effects in the heterozygous form become homozygous and express these effects on the components of fitness.
According to Charlesworth and Charlesworth (1987), there are two, not necessarily mutually exclusive, hypotheses that describe the decline in fitness with inbreeding; partial dominance and over-dominance. On the over-dominance hypothesis inbreeding depression results from the loss of advantage from the heterozygote state. This hypothesis assumes superiority of the heterozygous state, relative to the homozygous state. On the partial dominance hypothesis inbreeding decline results from the fixation in the homozygous state of recessive or partially recessive deleterious alleles.
It is possible that both of these mechanisms are in operation in cassava, with low germination rates and survival being indicative of the dominance hypothesis. The data generated in this study also indicated a general increase in mean performance for sprouting, vigour, height, flowering, dry matter content and harvest index among some partial inbred families at the CET which cannot be explained by over-dominance. When the surviving clones were evaluated for inbreeding depression (supplementary Tables 1 and 2), it was found that some families did not exhibit inbreeding depression for the evaluated fitness traits while others showed a low inbreeding depression, however it is likely that those individuals showing inbreeding depression did not survive, contributing to low survival rates. The low rate of survival also suggests that inbreeding depression is caused by genes of major effect or dominance (Ritland, 1996).
To our knowledge, this is the first study that has explored inbreeding for purposes of getting new resistance and/or higher resistance levels to CBSD. These findings are encouraging and thus justify the use of inbreeding in cassava, a highly heterozygous crop. Flowering which is critical in advancing generations of selfing appears not to be restrained by inbreeding in the clones used. This provides further motivation to explore inbreeding in cassava.
This study was initiated with a premise that inbreeding would significantly improve resistance to CBSD among partial inbreds as compared to their respective non-inbred progenitors. Results indicate that, within each family, a few S1 inbreds (1-15) showed higher levels of resistance than the S0 progenitors. It is, therefore, possible to get higher levels of resistance upon selfing. If field resistance is controlled by several heterozygous loci, it can be envisaged that more cycles of inbreeding for those clones that remained symptomless will lead to the generation of more new sources of resistance and/ or increase in levels of resistance to CBSD once all contributing loci are homozygous for the positive allele.
Alternatively, the generated S1 can be crossed in a different combination (between families) to exploit both additive and non-additive genetic effects of CBSD. Having molecular markers associated with these QTL would aid in the selection process.
The authors have not declared any conflict of interest.
This work was funded by International Institute of Tropical Agriculture through the project “Biotechnology tools to combat CBSD”. The authors thank the staff of NaCRRI that participated in the generation of partial inbreds and field data collection.
REFERENCES
|
Alicai T, Omongo CA, Maruthi MN, Hillocks RJ, Baguma Y, Kawuki R, Bua A, OtimNape GW, Colvin J (2007). Re-emergence of cassava brown streak disease in Uganda. Plant Dis. 91:24-29.
Crossref
|
|
|
|
Barrett SCH, Charlesworth D (1991). Effect of a change in the level of inbreeding on the genetic load. Nat. 352:522-524.
Crossref
|
|
|
|
|
Bigirimana S, Barumbanze P, Ndayihanzamaso P, Shirima R, Legg JP (2011). First report of cassava brown streak disease and associated Ugandan cassava brown streak virus in Burundi. New Dis. Rep. 24:26.
Crossref
|
|
|
|
|
Ceballos H, Iglesias CC, Pérez JC, Dixon AGO (2004). Cassava breeding: opportunities and challenges. Plant Mol. Biol. 56:503-515.
Crossref
|
|
|
|
|
Ceballos H, Fregene M, Pérez JC, Morante N, Calle F (2007). Cassava genetic improvement. In: M.S. Kang and P.M. Priyadarshan, editors, Breeding major food staples, pp 365-391 Blackwell Publishing, Ames, IA.
Crossref
|
|
|
|
|
Charlesworth D, Charlesworth B (1987). Inbreeding depression and its evolutionary consequences. Ann Rev. Ecol. Syst. 18:237-268. Charlesworth B (1992) Evolutionary rates in partially self-fertilizing species. Am. Nat. 140:126-148.
Crossref
|
|
|
|
|
Gondwe FMT, Mahungu NM, Hillocks RJ, Raya MD, Moyo CC, Soko MM, Chipungu FP, Benesi IRM (2003). Economic losses experienced by small-scale farmers in Malawi due to CBSVD. In: Legg J, Hillocks R (eds) Cassava Brown Streak Virus Disease: Past Present and Future. Proceedings of an International Workshop, Mombasa, Kenya, 27-30 October 2002. Natural Resources International Limited, Aylesford, UK. P 100.
|
|
|
|
|
Hallauer AR, Miranda FJB (1981). Quantitative genetics in maize breeding. Second Edition, 468 Iowa State Univ. Press. Iowa, USA.
|
|
|
|
|
Hillocks RJ, Jennings DL (2003). Cassava brown streak disease. A review of present knowledge and research needs. Int. J. Pest Manage. 49:225-234.
Crossref
|
|
|
|
|
Hull-Sanders MH, Eubanks MD (2005). Plant defense theory provides insights into interactions involving inbred plants and insect herbivores. Ecology 86(4):897-904.
Crossref
|
|
|
|
|
Jennings DL (1957). Further studies in breeding cassava for virus resistance. East Afr. Agric. J. 22:213-219.
Crossref
|
|
|
|
|
Kaweesi T, Kawuki R, Kyaligonza V, Baguma Y, Tusiime G, Ferguson ME (2014). Field evaluation of selected cassava genotypes for cassava brown streak disease based on symptom expression and virus load. Virol. J. 11:216.
Crossref
|
|
|
|
|
Kawuki RS, Nuwamanya E, Labuschagne MT, Herselman L, Ferguson M E (2011). Segregation of selected agronomic traits in six S1 cassava families. J. Plant Breed. Crop Sci. 3(8):154-160.
|
|
|
|
|
Kelly JK (1999a) Response to selection in partially self-fertilizing population. I. Selection on a single trait. Evolution 53:336-349.
Crossref
|
|
|
|
|
Kelly JK (1999b) Response to selection in partially self-fertilizing population. II. Selection on multiple traits. Evolution 53:350-357.
Crossref
|
|
|
|
|
Kulembeka HP, Ferguson M, Herselman L, Kanju E, Mkamilo G, Masumba E, Fregene M, Labuschagne MT (2012) Diallel analysis of field resistance to brown streak disease in cassava (Manihot esculenta Crantz) landraces from Tanzania: Euphytica 187:277-288.
Crossref
|
|
|
|
|
Mbanzibwa DR, Tian YP, Tugume AK, Mukasa SB, Tairo F, Kyamanywa S, Kullaya, A, Valkonen JPT (2011). Simultaneous virus-specific detection of the two cassava brown streak associated viruses by RT-PCR reveals wide distribution in East Africa, mixed infections, and infections in Manihot glaziovii. J. Virol. Methods 171:394-400.
Crossref
|
|
|
|
|
Mbewe W, Kumar PL, Changadeya W, Ntawuruhunga P, Legg J (2015). Diversity, distribution and effects on cassava cultivars of cassava brown streak viruses in Malawi. J. Phytopathol. 163(6):433-443.
Crossref
|
|
|
|
|
Monger WA, Seal S, Cotton S, Foster GA (2001). Identification of different isolates of cassava brown streak virus and development of a diagnostic test. Plant Pathol. 50:768-775.
Crossref
|
|
|
|
|
Mulimbi W, Phemba X, Assumani B, Kasereka P, Muyisa S, Ugentho H, Reeder R, Legg J, Laurenson L, Weekes R (2012). First report of Ugandan cassava brown streak virus on cassava in Democratic Republic of Congo. New Dis. Rep. 26:2044.
Crossref
|
|
|
|
|
Munga TL (2008). Breeding for cassava brown streak resistance in coastal Kenya. PhD Thesis, University of KwaZulu-Natal, South Africa.
|
|
|
|
|
Ndunguru J, Sseruwagi P, Tairo F, Stomeo F, Maina S, Djikeng A, Kehoe M, Boykin LM (2015). Analyses of twelve new whole genome sequences of cassava brown streak viruses and Ugandan cassava brown streak viruses from East Africa: diversity, supercomputing and evidence for further speciation. Plos One 10 (10).
|
|
|
|
|
Payne RW, Harding SA, Murray DA, Soutar DM, Baird DB, Glaser AI, Welham SJ, Gilmour AR, Thompson R, Webster R (2011). The guide to Genstat release 14, part 2: statistics. VSN International, Hemel Hempstead, UK.
|
|
|
|
|
Pennisi E (2010). Armed and dangerous. Science 327:804-805.
Crossref
|
|
|
|
|
Ritland K (1996). Inferring the genetic basis of inbreeding depression in plants. Genome 39:1-8.
Crossref
|
|
|
|
|
Tumuhimbise R, Melis R, Shanahan P (2014). Diallel analysis of early storage root yield and disease resistance traits in cassava (Manihot esculenta Crantz) Field Crop Res. 167:86-93.
Crossref
|
|
|
|
|
Tomlinson JA, Ostoja-Starzewska S, Adams IP, Miano DW, Abidrabo P, Kinyua Z, Alicai T, Dickinson MJ, Peters D, Boonham N, Smith J (2013). Loop-mediated isothermal amplification for rapid detection of the causal agents of cassava brown streak disease. J. Virol. Methods 191(2):148-154.
Crossref
|
|
|
|
|
Walsh B (2005). The struggle to exploit non-additive variation. Aust. J. Agric. 56:873-881.
Crossref
|
|
|
|
|
Zacarias AM, Labuschagne MT (2010). Diallel analysis of cassava brown streak disease, yield and yield related characteristics in Mozambique: Euphytica 176:309-320.
Crossref
|
|