ABSTRACT
Viral infections are a major challenge to sustainable cowpea production in sub-Saharan Africa (SSA). Nine cowpea (Vigna unguiculata L. Walp) genotypes were evaluated for resistance against viral infection in a field trial involving randomized complete block design with 4 replications. Viral disease severity was assessed at 2, 4 and 6 weeks after planting (WAP) based on disease symptoms. Double antibody sandwich-enzyme linked immunosorbent assay (DAS-ELISA) using antisera raised against cucumber mosaic virus (CMV), cowpea severe mottle virus (CPSMV), cowpea mosaic virus (CPMV) and southern bean mosaic virus (SBMV) was used to detect the viruses associated with diseased leaf samples collected from the field. Biometric and yield data were also taken. The mean disease incidence and area under disease progress curve (AUDPC) varied significantly (P < 0.05) among the cowpea genotypes, with 7 (Apagbaala, UCC-366, UCC-489, UCC-490, UCC-497, UCC-514 and UCC-523) out of 9 cowpea genotypes showing field resistance whilst the other two (UCC-473 and UCC-484) exhibited moderate resistance. ELISA showed that all the 9 cowpea genotypes were infected with at least one of the three viruses namely CMV, CPMV and CPSMV, whereas SBMV was not detected. Co-infection by CMV, CPMV and CPSMV was observed in UCC-366. Mean plant height, canopy diameter and seed yield differed significantly (P < 0.05) among the cowpeas. UCC-473, UCC-316, and UCC-523 had high mean seed yields of 6.690, 4.922 and 4.144 t ha-1 respectively, above the overall mean seed yield of 3.63 t ha-1, emphasizing their resilient to viruses.
Key words: Vigna unguiculata, viral infection, disease incidence and severity, resistance screening.
Legumes play important roles in provision of food security, generation of income, and maintenance of environment in most smallholder farming systems in sub-Saharan Africa (SSA) (Odendo et al., 2011). Africa produces about 8 million tonnes of grain legumes estimated at about 70% of the total global production, from 17.7 million hectares of land (IITA, 2007).Cowpea (Vigna unguiculata L.Walp) is a major staple food crop in
sub-Saharan Africa (SSA), especially in the dry savanna regions of West Africa (Asare et al., 2010; Dugje et al., 2009).The seeds are a major source of plant proteins and vitamins for man, feed for animals, and also a source of cash income. Cowpea leaves and green pods are consumed as vegetable and the dried grain is used in many different food preparations (Dugje et al., 2009; Kyei-Boahen et al., 2017).
Cowpea is an essential component of the cropping systems because it fixes atmospheric nitrogen and contributes to soil fertility improvement particularly in smallholder farming systems where little or no fertilizer is used (Kyei-Boahen et al., 2017). The crop is drought tolerant and adapted to stressful environments (Bisikwa et al., 2014; Ddamulira et al., 2015) such as the prevailing conditions of the dry savannah regions in Ghana. Cowpea is an important food security crop and a major source of income especially in the Northern and Volta regions of Ghana where the bulk of the crop is produced.
Viruses are a major biotic constraint to cowpea production, reported to bring about yield losses ranging from 10 to 100% (Dhanasekar and Reddy, 2015). Over 140 viruses worldwide have been reported to attack cowpea and at least 11 of these occur in Africa (Hughes and Odu, 2003). Viruses considered most damaging to cowpea in Africa are bean common mosaic virus-black eye cowpea mosaic (BCMV-BICM), cowpea aphid-borne mosaic virus (CABMV), cucumber mosaic virus (CMV), cowpea mosaic virus (CPMV), cowpea severe mosaic virus (CPSMV), southern bean mosaic virus (SBMV) and cowpea mottle virus (CPMoV) (Hampton et al., 1997). Others are cowpea mild mottle virus (CPMMV) (Jeyanandarajah and Brunt, 1993), cowpea golden mosaic geminivirus (CGMV) and cowpea chlorotic mottle virus (CCMV) (Hampton et al., 1997). Viruses so far reported to be infecting cowpea in Ghana include SBMV, CABMV, BICMV and cowpea mild mottle virus (CPMMV) (Jeyanandarajah and Brunt, 1993; Lamptey and Hamilton, 1974; Zettler and Evans, 1972). The seed-borne nature of these viruses renders them very destructive to emerging seedlings and insect vectors can spread these further (Ndiaye et al., 1993; Bashir et al., 2002).
Effective management of these viruses is important to improve yields and quality of cowpea. Managing these viruses with insecticides is not effective because they are transmitted by several insect species in a non-persistent manner (Umaharan et al., 1997). The most economical, practicable and effective method of reducing crop losses due to viral infection is through the use of resistant varieties (Taiwo, 2003; Mbeyagala et al., 2014). Development of resistant varieties against different type and strain of viruses entails screening of germplasm in a particular agro-climate for identification of resistance to the particular strain prevailing in that region. However, field screening for virus resistance based solely on symptoms is not reliable (Shoyinka et al., 1997), as different viruses display overlapping symptoms (Dhanasekar and Reddy, 2015). Moreover, plants can also exhibit virus-like symptoms when exposed to adverse weather conditions, soil nutrient imbalances, pest infestations and non-viral infections (Hughes and Odu, 2003). Nine out of thirty-two (32) cowpea genotypes that were screened against viral infection under natural conditions in Ghana during 2015 major cropping season (Essandoh et al., 2017) were observed to exhibit mild symptoms of viral infections, indicating field resistance. Viruses associated with these symptoms are unknown and the performance of these resistant genotypes under different environment is also unknown. This study was therefore conducted to evaluate the performance of nine cowpea genotypes and to identify the associated viruses.
Study area
Field experiment was conducted at the Teaching and Research farm of the School of Agriculture, University of Cape Coast (UCC), which falls within the coastal savannah agro-ecological zone of Ghana, from July to October 2016 during the major cropping season. This location (5°10’N, 1.2°50’W) has Acrisol soil type and a distinct bimodal rainfall behaviour, with a major rainy season (May-June) and a minor rainy season (August - October) with an annual rainfall ranging from 750 to 1000 mm (Parker et al., 2010). Temperatures of the area range from 23.2 to 33.2ºC with an annual mean of 27.6°C (Owusu-Sekyere et al., 2011).
Plant material
A total of nine cowpea genotypes were used for the study. These included eight recombinant inbred lines (F1, F2 or F3 generations) from University of Cape Coast (UCC-366, UCC-473, UCC-484, UCC-489, UCC-490, UCC-497, UCC-514, UCC-523) and the genotype Apagbaala from Savannah Agricultural Research Institute (SARI), Nyankpala, Ghana.
Experimental design and field layout
The randomized complete block design with nine treatments (cowpea genotypes) and four replications was used. A total land area of 2880 m2 measuring 40 m x 72 m was ploughed and harrowed to render the soil loose. It was then divided into four blocks, spaced 1.5 m apart, and each block was further divided into 9 plots, spaced 1 m apart, and a plot size of 4 m x 4 m. Three seeds were sown per hill with an inter row spacing of 40 cm and intra-row spacing of 60 cm and later thinned to two plants per hill. There were four rows per plot, with 17 plants in a row, making 68 plants per plot.
Cultural practices
The study was under rain-fed conditions. Weeding was done manually using a machete and a hoe, as well as pulling out weeds by hands, when necessary. Fertilizer (NPK 15:15:15) was applied at a rate of 250 kg ha-1.
Data collection
Data was collected on disease incidence and severity, plant height, canopy diameter and seed yield. In each case data was taken from 10 inner rows of each plot and the mean per plant determined. Data on disease incidence and severity were assessed at 2, 4 and 6 weeks after planting (WAP), based on disease symptoms described by Gumedzoe et al. (1998).
Incidence of virus disease for the various fields was calculated using the formula:
The severity of viral disease in each field was assessed based on the 1 to 5 symptom severity scale developed by Gumedzoe et al. (1998) as shown in Table 1.
Detection of viruses from cowpea samples
Symptomatic leaf samples were collected from six weeks old plants of each of the eight cowpea genotypes from the field in order to identify viruses associated with them. Between three and ï¬ve young leaves were taken from each plant sampled. Virus identification was done using standard double antibody sandwiched enzyme-linked immunosorbent assay (DAS-ELISA) as described by Clark and Adams (1977) using polyclonal antisera raised against CPMV, CPSMV, CMV and SBMV (DSMZ, Braunschweig, Germany).
The leaf samples were pulverized with mortar and pestle in an extraction buffer (8.0 g NaCl, 0.2 g KH2PO4, 1.1 g Na2HPO4, 0.2 g KCl/L, pH 7.4) containing 0.05% v/v Tween 20, and 2% w/v polyvinylpyrolidone at a 1:10 ratio (tissue weight: extraction buffer volume). The microtitre plates (96 wells Nunc, Maxisorp, Roskide, Dernmark) were coated with primary (coating) antibody (IgG, 1/200 in coating buffer, Na2CO3, NaHCO3, NaN2) and incubated at 37°C for 4 h. After washing the plates four times with phosphate-buffer saline pH 7 with Tween 20 (PBS-T), the wells were loaded with the leaf extracts at 200 µL extract per well and incubated overnight (for 18 h) at 4°C.The plates were then washed four times with PBS-T, and incubated with enzyme conjugate (alkaline phosphatase conjugate (Sigma-Aldrich, Inc.), diluted at 1/200 in PBS-T+BSA+NaN2) at 37°C for 2 h. After washing the plates four times with PBS-T, they were incubated for 1 hour at room temperature with freshly prepared phosphate substrate solution (100 µL per well) composed of p-nitrophenyl phosphate tablet (ADGEN Phytodiagnostics) at 1.0 mg mL-1 in 9.87% diethanolamine, pH 9.8. Healthy leaf sample and purified individual virus samples from DSMZ were included as negative and positive controls respectively in order to test the specificity and efficiency of the various polyclonal antibodies used in the assay.
The absorbance values at 405 nm (A405) were recorded using Anthos microplate reader (Biochrum Ltd, Cambridge, UK). Absorbance values of three uninfected leaf samples were also measured. A test sample was deemed to be positive when the A405 was higher than 3 times the mean absorbance of the uninfected leaf samples (threshold value).
Data analysis
Data on mean viral disease severity scores were used to calculate area under the disease progress curve (AUDPC) for each of the cowpea genotypes according to Shaner and Finney (1977):
where: Yi = disease severity at the ith observation; Xi = time (weeks) at the ith observation, and n = total number of observations
The AUDPC, which is a quantitative summary of disease intensity over a period 10 weeks was used to measure disease resistance in each cowpea genotype. Data on disease incidence was arcsine-transformed in order to homogenize the variances before subjecting to analyses of variance (ANOVA). The other data (disease incidence and severity, AUDPC, plant height, canopy diameter, and yield) were also subjected to ANOVA and the mean separated by least significance difference (LSD) method at 5% level of probability. Pearson's correlation coefficients were calculated for the relationships between disease, biometric and seeds yield data. All statistical analyses were performed using GenStat Release version 12 (VSN International).
Viral symptoms observed on the field
Varying degrees of symptoms were observed on the field (Figures 1a to e). These include stunted growth, yellow mosaic, vein clearing, mottling of leaves and necrotic lesions.
Incidence and severity of cowpea viruses on selected cowpea genotypes
Viral disease incidence showed significant differences among the cowpea genotypes at 2 WAP (F8, 24 = 43.08; P<0.001), 4 WAP (F8, 24 = 6.91; P<0.001) and 6 WAP (F8, 24 = 7.02; P< 0.001) (Table 2). At 2 WAP, the genotype UCC-484 had the highest mean disease incidence (90%) which was significantly different (P<0.05) from all the other cowpea genotypes. On the other hand, the genotypes, ApagbaalaUCC-514 and UCC-489 had mean viral incidence of 0% each which was not significantly different (P > 0.05) from the mean incidence of 7.24% recorded for the genotype UCC-366 (Table 2).
At 4 WAP, the genotype UCC-473 had the highest mean disease incidence of 68.58% while Apagbaala had the lowest mean disease incidence of 7.24% (Table 2). At 6 WAP, the genotype UC-96-473 had the highest mean disease incidence (68.58%) while Apagbaala had 0%. Mean disease incidence declined between 4 and 6 WAP in Apagbaala, UCC-489 and UCC-497 (Table 2).
The result also revealed that the mean final disease severity scores recorded for the cowpea genotypes varied significantly (P < 0.05) among them (F8, 24 = 102.72; P<0.001) (Table 2). Apagbaala displayed the lowest mean disease severity score (1.00) which was not significantly different (P > 0.05) from the genotypes UCC-514 and UCC-489 with mean severity scores of 1.03 and 1.07 respectively but was significantly different (P < 0.05) from the remaining 6 genotypes. Conversely, genotype UCC-484 had the highest mean final severity score of 2.00, which was significantly different (P<0.05) from the other genotypes.
The area under disease progress curve (AUDPC) showed significant difference among the cowpea genotypes (F8, 24=168.16; P<0.001) as shown in Table 2. Genotype UCC-484 had the highest AUDPC of 7.87 which was significantly different from all the other genotypes, indicating that UCCC-484 accumulated the highest disease pressure during the entire growing period. On the other hand, the genotype Apagbaala had the lowest AUDPC score of 4.07 which was not significantly different (P > 0.05) from the genotypes UCC-489 and UCC-514 with AUDPC of 4.20 and 4.10 respectively, indicating that they experienced the lowest disease pressure during the entire growing period.
Biometric characters and seeds yield
ANOVA showed significant differences among the cowpea genotypes in respect of their mean plant heights (F8, 24 = 20.86; P<0.001), mean canopy diameters (F8, 24 = 4.36; P<0.002) and mean seed yields (F8, 24 = 2.98; P<0.018) as shown in Table 3. The highest mean plant height (41.84 cm) was recorded for the genotype UCC-366 which was not significantly different (P > 0.05) from the genotype UCC-484 with a mean plant height of 36.96 cm. Genotype UCC-473 had the lowest plant height (13.03 cm) which was not significantly different (P > 0.05) from that of the genotypes, UCC-490 and Apagbaala with a mean plant height of 17.77 and 14.71 cm, respectively. Similarly, the highest canopy diameter (150.1 cm) was recorded for genotype UCC-366 which was not significantly different (P > 0.05) from the genotypes UCC-484, UCC-490, UCC-497 and UCC-523 with mean canopy diameters of 127.6, 122.7, 128.3 and 133.4 cm, respectively. Also, genotype UCC-473 had the lowest canopy diameter (67.1 cm) which was not significantly different (P > 0.05) from the genotypes, Apagbaala, UCC-489 and UCC-514 with mean canopy diameters of 88.5, 97.4 and 102.6 cm, respectively (Table 3).
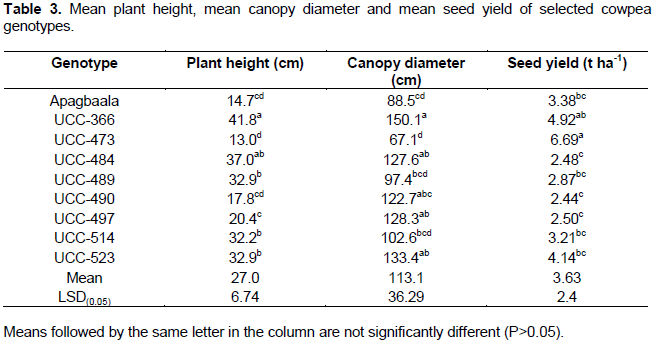
Genotype UCC-473 had the highest seed yield (6.690 t ha-1) which was not significantly different (P>0.05) from the genotype UCC-366 with a mean seed yield of 4.922 t ha-1 but significantly higher (P<0.05) from the other genotypes. The seed yield recorded for UCC-366 was not significantly different (P<0.05) from the genotypes UCC-523, Apagbaala, UCC-514 and UCC-489 with mean seed yields of 4.144, 3.379, 3.208 and 2.87 t ha-1, respectively (Table 3).
Correlations among the variables
Correlation coefficients among the variables studied are shown in Table 4. AUDPC is correlated significantly positive with initial incidence (r=0.9123; P<0.01) and initial severity score (r=0.9708; P<0.01) (Table 4).There was also a highly significant positive correlation between initial incidence and initial severity (r=0.9040; P<0.01) (Table 4). Further, there was a significant positive correlation between plant height and canopy diameter (r=0.4317; P<0.05).
Viral detection by double antibody sandwiched enzyme-linked immunosorbent assay (DAS-ELISA)
Three virus species namely CPMV, CPSMV and CMV were detected in the cowpea genotypes using DAS-ELISA and each sample was infected with at least one of the three viruses (Table 5). It was observed that some viruses were associated with single or multiple infection(s) in the plant samples. The leaf samples had a high prevalence of single virus infection compared with multiple virus infections. In single virus-infected leaf samples, CMV was the most prevalent, infecting 87.7% of the samples tested, followed by CPSMV which infected 25% whereas CPMV was the least prevalent, infecting only 12.5%. Co-infection by CMV + CPSMV + CMV was observed in one cowpea genotype UCC-366. SBMV was not detected in any of the plant samples tested.
The virus symptoms observed on cowpea on the field
Symptoms observed in the field (mosaic, leaf mottling, vein clearing and necrotic lesion) were similar to symptoms reported elsewhere on legumes affected by viral diseases (Akinjogunla, 2005; Aliyu et al., 2012). The varying symptoms observed in the study are suggestive of different viruses which infect cowpeas in the study area (Aliyu et al., 2012). The variation in symptoms observed in the study could be attributed to the type of viral strains infecting the cowpea, cowpea cultivar, the time of infection, light intensity, environmental temperature, and mixed infections (Jones et al., 1991; Aliyu et al., 2012).
Incidence and severity of viruses on selected cowpea genotypes
The study to evaluate nine cowpea genotypes for resistance to viral infections under natural conditions showed variation in the disease incidence and severity among the cowpea genotypes at various sampling dates (WAP). Variation in the level of viral infection among the cowpea genotypes in the current study could be related to their genetic variability. This finding is in agreement with the work done by Orawu (2007)in his study on the occurrence of CABMV and prospects of improving resistance in local cowpea landraces in Uganda. Ojuederie et al. (2009)also reported that the reaction of various cowpea accessions to viral disease is genotype dependent. Variation in the incidence and severity of viral disease among the cowpea genotypes could also be due to the age of the plants at the time of infection. According to Picó et al. (1996), plants infected or inoculated at older age produce milder symptoms which may be wrongly considered as manifestation of genetic resistance. This corroborates the report of Ehinmore and Kareem (2010)which states that infection at a later stage results in reduced effects because at that stage, the plants are more mature and the virus has little or no deleterious effects on them. Lapidot (2007)also reported that the success of TYLCV infection of beans is highly dependent on the bean plant age.
With the exception of UCC-473 and UCC-484, which exhibited moderate resistance, the other cowpea genotypes exhibited mild symptoms of viral infection (Table 2). In this study, genotypes Apagbaala, UCC-366, UCC-489, UCC-490, UCC-497, UCC-514 and UCC-523 with low disease severity in terms of AUDPC and low final severity scores may offer single or multiple virus resistance, which is comparable to previous work (Essandoh et al., 2017), where these nine cowpea genotypes were found to exhibit field resistance. This finding also corroborates the report by Mbeyagala et al. (2014) when they screened 105 cowpea genotypes for resistance against viral infection under natural condition in Uganda. The ELISA serology revealed that genotype UCC-366 that also exhibited field resistance was co-infected with CMV, CPSMV and CMV (Table 5), demonstrating multiple field resistance against these three viral species. On the other hand, the ELISA detected single viral species from the other genotypes (UCC-489, UCC-490, UCC-497, UCC-514 and UCC-523) suggesting that they offered single virus resistance.
The differences in the levels of prevalence of CMV, CPSMV and CMV in the cowpea genotypes could be explained on the basis of antagonism, inoculum level, the age of the plant, climatic conditions and cultivar type (Orawu et al., 2015). The varying reactions of the cowpea genotypes to different cowpea viruses observed in the present study is quite valuable because it will enable breeders to breed for resistance against viruses that prevail at a particular location, as reported by Dhanasekar and Reddy (2015).
The presence of viruses in a combination may result in synergism or antagonism effects within the infected plants. For instance, viruses acting in a synergistic manner enhance their infection rate, thus leading to the development of complexes of diseases (Syller and Grupa, 2016; Syller, 2012; Fondong et al., 2000). When viruses are antagonised when in a combination with other viruses, their rate of infection may be affected compared with single virus infection (Syller and Grupa, 2016; Syller, 2012). In the present study, genotype UC-366 was co-infected with CMV, CPSMV and CMV yet it exhibited mild symptom severity and low AUDPC compared to genotypes UCC-473 and UCC-484 which were infected with only CMV. This suggests that the three viruses (CMV, CPSMV and CMV) which infected UC-366 exhibited antagonism among them, in contrast with synergism as reported (Syller, 2012; Fondong et al., 2000). Nonetheless, it has been reported (Syller, 2012) that multiple infections may result in the generation of variants showing novel genetic features, and thus change the genetic structure of the viral population. Hence, understanding the interactions between CMV, CPSMV and CMV in cowpea may be of crucial significance for the understanding of viral pathogenesis and evolution, and consequently for the development of efficient and stable management strategies, as suggested by Syller (2012).
There were variations in the prevalence of the three viral species (CMV, CPSMV and CMV) detected by ELISA from the cowpea genotypes. CMV was found to be the most prevalent virus species in the study area, infecting 88.9% of the cowpea genotypes compared to CPSMV and CPMV and which infected only 22.2 and 11.1% of the cowpea genotypes respectively. The high infection rate of CMV observed in the current study could be due to the fact that it is highly seed borne in many cowpea varieties as reported by Thottappilly and Rossel (1987). The finding of this study also agrees with the report of Van Regenmortel et al. (2000) which states that Bromoviridae including CMV, is one of the most important widespread viruses in the world infecting the largest number (approximately 1000) of plant species. The higher infection rate of the cowpea genotypes by CMV compared to CPMV and CPSMV could suggest its relative persistence under adverse environmental conditions over the other viruses.
Leaf samples for all the accessions tested negative for SBMV, although the samples were symptomatic. Similar result was obtained by Ojuederie et al. (2009)in their study of serological detection of seed borne viruses in cowpea regenerated germplasm using protein A sandwich-ELISA. This could be due to low virus concentrations in the leaf samples or the presence of serologically variable strains of the viruses and the non-availability of antibodies specific to them (Aliyu et al., 2012).
Mean disease incidence declined between 4 and 6WAP in Apagbaala, UCC-489 and UCC-497, suggest symptom recovery (Table 2). In the case of Apagbaala, there was total recovery with the emergence of asymptomatic leaves following a systemic infection. According to Goshal and Sanfacon (2015), symptom is generally accompanied with reduced virus titres and sequence specific resistance to secondary infection. Jovel et al. (2007) had earlier argued that induction of recovery does not require a reduction of virus titre, and suggest that viral proteins RNAs or virus derived siRNA function to counteract host defense responses.
Average growth and yield performance of selected cowpea genotypes
Significant variations in the growth and yield among the cowpea genotypes (Table 3) could be due to different host-virus interactions (Anneke et al., 2013), age of plants at which plants were infected (Taiwo et al., 2007; Sastry and Singh, 1974) and genetic constitution of the cowpea genotypes. Taiwo et al. (2007) reported that viral infection of cowpea at early age results in more severe symptoms, sometimes resulting in death of the affected plants. This may explain why the genotype UCC-484 which experienced early and 90% disease incidence had the highest severity score), highest AUDPC and had a low seed yield (2.482 t ha-1) (Table 3). Significant negative correlation between AUDPC and seed yield in the present study (Table 4) further indicates that as the disease pressure increases the yield of the plant reduces. Similar result was obtained by Orawu (2007)when he evaluated cowpea genotypes for resistance to CABMV infection in Uganda. This observation could be due to the fact that the cowpea plants infected by viruses activate sophiscated defense pathways which operate at different levels, often at significant fitness costs, resulting in yield reduction as reported by Syller and Grupa (2016). Similarly, in maize-maize dwarf mosaic virus (MDMV) pathosystem, it was reported that the virus infection in maize induce necrotic disease which results in up to 91% yield loss and death of many plants especially when infection occur early( Uyemoto et al., 1981).
Although genotype UCC-473 that displayed early viral symptom, and had moderately severe infection and high AUDPC, it had a seed yield of 6.69 t ha-1 which was higher than the seed yield for all the cowpea genotypes (3.63 t ha-1). This observation could be due to the plant’s ability to tolerate viral infection or recover from the damage by the disease (Kessler and Baldwin, 2002; Teetes, 1996)or an infection by mild strain of the virus.
Generally, genotypes UCC-473, followed by UCC-316, and UCC-523 had mean seed yields of 6.690, 4.922 and 4.144 t ha-1 respectively, above the mean seed yield of 3.63 t ha-1 for all the nine cowpea genotypes.
The study identified 7 (Apagbaala, UCC-366, UCC-489, UCC-490, UCC-497, UCC-514 and UCC-523) out of 9 cowpea genotypes showing field resistance whilst the other two (UCC-473 and UCC-484) exhibited moderate resistance, in respect of their AUDPC values and final severity scores. ELISA serology revealed that each of the nine cowpea genotypes was infected with at least one of the three viral species namely CMV, CPSMV and CPMV, suggesting none was immune to virus infection. CMV was found to be the most prevalent virus species in the study area, infecting eight (8) out of nine (9) cowpea genotypes compared to CPSMV and CPMV which infected two and one cowpea genotypes, respectively. Genotype UCC-473, followed by genotypes UCC-316 and UCC-523 had mean seed yields of 6.690, 4.922 and 4.144 t ha-1, respectively, above the mean seed yield of 3.63 t ha-1 for all the nine cowpea genotypes. Multi-locational evaluation of these three cowpea genotypes could be carried out prior to their release as varieties.
The authors have not declared any conflict of interests.
REFERENCES
|
Aliyu TH, Balogun OS, Kumar L (2012). Survey of the Symptoms and Viruses Associated with Cowpea (Vigna Unguiculata (L).) in the Agroecological Zones of Kwara State, Nigeria. Ethiop. J. Environ. Stud. Manage. 5(4):613-619
Crossref
|
|
|
|
Anneke E, Hogerwerf L, Slingenbergh J (2013). Pathogen-host environment interplay and disease emergence. Emerg. Microbes Infect. 2:1-7.
|
|
|
|
|
Asare A, Gowda B, Galyuon I, Aboagye L, Takrama J, Timko M (2010). Assessment of the genetic diversity in cowpea (Vigna unguiculata L. Walp.) germplasm from Ghana using simple sequence repeat markers. Plant Geneti. Resour. 8(2):142-150.
|
|
|
|
|
Bashir M, Ahmad Z, Ghafor A (2002). Cowpea aphi-borne mosaic potyvirus: A review, Int. J. Pest Manage. 48(2):155-168.
Crossref
|
|
|
|
|
Bisikwa J, Kawooya R, Ssebuliba JM, Ddungu SP, Biruma M, Okello DK (2014). Effects of plant density on the performance of local and elite cowpea varieties in Eastern Uganda. Afr. J. Appl. Agric. Sci. Technol. 1:28-41.
|
|
|
|
|
Ddamulira G, Santos CAF, Obuo P, Alanyo M, Lwanga CK (2015). Grain yield and protein content of Brazilian cowpea genotypes under diverse Ugandan environments. Am. J. Plant Sci. 6:2074=2084.
Crossref
|
|
|
|
|
Dhanasekar P, Reddy K (2015). Serological screening of cowpea genotypes for resistance against cowpea aphid borne mosaic virus using DAS-ELISA. Asian J. Plant Pathol. 9:83-90.
Crossref
|
|
|
|
|
Dugje IY, Omoigui LO, Ekeleme F, Kamara AY, Ajeigbe H (2009). Farmers' guide to cowpea production in West Africa. Ibadan, Nigeria. International Institute of Tropical Agriculture (IITA), 2009.
|
|
|
|
|
Ehinmore I, Kareem KT (2010). Effect of amaranthus mosaic virus on the growth characters of Amaranthus hybridus.ABJNA. Agric. Biol. J North Am. 1(2):75-79.
|
|
|
|
|
Essandoh AV, Asare-Bediako E, Asare TA, Kusi F, Aboagye ML (2017). Multi-locational screening of genotypes of cowpea (Vigna unguiculata L. Walp) for resistance to viral infection. Ann. Res. Rev. Biol. 14(4):1-13.
Crossref
|
|
|
|
|
Fondong VN, Pita JS, Rey MEC, De Kochko A, Beachy RN, Fauquet CM (2000). Evidence of synergism between African cassava mosaic virus and a new double-recombinant geminivirus infecting cassava in Cameroon. J. Gen. Virol. 81(1):287-297.
Crossref
|
|
|
|
|
Goshal B, Sanfacon H (2015). Symptom recovery in virus-infected plants: revisiting the role of RNA silencing mechanisms. Virol. 479-480.
Crossref
|
|
|
|
|
Gumedzoe MYD, Rossel HW, Thottappilly G, Asselin A, Huguenot C (1998). Reaction of cowpea (Vigna unguiculata L. Walp.) to six isolates of blackeye cowpea mosaic virus (BlCMV) and cowpea aphid-borne mosaic virus (CAMV), two potyviruses infecting cowpea in Nigeria. Int. J. Pest Manage. 44(1):11-16.
Crossref
|
|
|
|
|
Hampton RO, Thottappilly G, Rossel HW (1997). Viral diseases of cowpea and their control by resistance-conferring genes. Adv. Cowpea Res. pp. 159-175.
|
|
|
|
|
Hughes JDA, Odu BO (2003). Plant Virology in Sub-Saharan Africa: Proceedings of a Conference Organized by IITA: 4-8 June 2001, International Institute of Tropical Agriculture, Ibadan, Nigeria: IITA.
|
|
|
|
|
International Institute of Tropical Agriculture (IITA) (2007).Cowpea.IITAannualreport.
View
|
|
|
|
|
Jeyanandarajah P, Brunt AA (1993). The natural occurrence, transmission, properties and possible affinities of cowpea mild mottle virus. J. Phytopathol. 137(2):148-156.
Crossref
|
|
|
|
|
Jovel J, Walker M, Sanfacon H (2007). Recovery of Nicotiana benthamiana plants from a necroticc response induced by Nepovirus is associated with RNA silencing but not with reduced virus titre. J. Virol. 81(22):12285-12297.
Crossref
|
|
|
|
|
Kessler A, Baldwin IT (2002). Plant responses to insect herbivory: the emerging molecular analysis. Ann. Rev. Plant Biol. 53(1):299-328.
Crossref
|
|
|
|
|
Kyei-Boahen S, Canon EN, Savala CEN, David Chikoye D, Robert Abaidoo R (2017).Growth and yield responses of cowpea to inoculation and phosphorus fertilization in different environments. Front. Plant Sci. 8:646-659.
Crossref
|
|
|
|
|
Lamptey PNL, Hamilton RI (1974). A new cowpea strain of southern bean mosaic virus from Ghana. Phytopathol. 64:1100-1104.
Crossref
|
|
|
|
|
Lapidot M (2007). Screening for TYLCV-resistance plants using whitefly-mediated inoculation tomato yellow leaf curl virus disease. Springer. pp. 329-342.
Crossref
|
|
|
|
|
Mbeyagala EK, Mukasa BS, Tukamuhabwa F, Bisikwa J (2014). Evaluation of cowpea genotypes for viral resistance under natural conditions in Uganda. J. Agric. Sci. 6(10):176-187.
|
|
|
|
|
Ndiaye M, Bashir M, Keller K, and Hampton RO (1993). Cowpea viruses in Senegal, west Africa: identities, distribution, seed transmission and sources of resistance. Plant Dis. 77:999-1003.
Crossref
|
|
|
|
|
Odendo M, Bationo A, Kimani S (2011). Socio-Economic Contribution of Legumes to Livelihoods in Sub-Saharan Africa. In: Bationo A., Waswa B., Okeyo J., Maina F., Kihara J., Mokwunye U. (eds) Fighting Poverty in Sub-Saharan Africa: The Multiple Roles of Legumes in Integrated Soil Fertility Management. Springer, Dordrecht.
Crossref
|
|
|
|
|
Ojuederie OB, Odu BO, Ilori CO (2009). Serological detection of seed borne viruses in cowpea regenerated germplasm using protein a sandwich enzyme linked immunorsorbent assay. Afr. Crop Sci. J. 17(3):125 - 132.
|
|
|
|
|
Orawu M (2007). Occurrence of cowpea aphid-borne mosaic virus and prospects of improving resistance in local cowpea landraces in Uganda. (Doctorial thesis), University of KwaZulu-Natal. Republic of South Africa.
|
|
|
|
|
Orawu M, Obuo J, Omadi R (2015). Distribution and Detection of Cowpea Viruses Infecting Cowpea in Uganda. Am. J. Plant Sci. 6:574-581.
Crossref
|
|
|
|
|
Owusu-Sekyere JD, Alhassan M, Nyarko BK (2011). Assessment of climate shift and crop yields in the Cape Coast area in the Central Region of Ghana. ARPN J. Agric. Biol. Sci. 6(2):49-54.
|
|
|
|
|
Parker BQ, Osei BA, Armah FA, Yawson DO (2010). Impact of biomass burning on soil organic carbon and the release of carbon dioxide into the atmosphere in the coastal savanna ecosystem of Ghana. J. Renew Sustain. Energy 2(3):1-7.
Crossref
|
|
|
|
|
Picó B, Díez MJ, Nuez F (1996). Viral diseases causing the greatest economic losses to the tomato crop. II. The tomato yellow leaf curl virus- A review. SciHortic. 67(3-4):151-196.
Crossref
|
|
|
|
|
Sastry KSM, Singh SJ(1974). Effect of yellow vein mosaic virus infection on growth and yield of okra crop. Ind. Phytopathol. 27(3):294-297.
|
|
|
|
|
Shaner G, Finney RE (1977). The effect of nitrogen fertilization on the expression of slow vmildewing resistance in Knox wheat.Phytopathol. 57:1051-1056.
Crossref
|
|
|
|
|
Shoyinka SA, Thottappilly G, Adebayo GG, Anno-Nyako FO (1997). Survey on cowpea virus incidence and distribution in Nigeria. Int. J. Pest Manage. 43(2):127-132.
Crossref
|
|
|
|
|
Syller J, Grupa A (2016). Antagonistic within-host interactions between plant viruses: molecular basis and impact on viral and host fitness. Mol. Plant Pathol. 17(5):769-782.
Crossref
|
|
|
|
|
Syller J (2012). Facilitative and antagonistic interactions between plant viruses in mixed infections. Mol. Plant Pathol. 13(2):204-216.
Crossref
|
|
|
|
|
Taiwo MA, Kareem KT, Insa IY, D'A Hughes J (2007). Cowpea viruses: Effect of single and mixed infections on symptomatology and virus concentration. Virol. J. 4:95-99.
Crossref
|
|
|
|
|
Taiwo MA (2003). Viruses infecting legumes in Nigeria: case history. Plant Virology in Sub-Saharan Africa, Hughes, JA and BO Odu (Eds.). IITA, Nigeria. pp. 365-380.
|
|
|
|
|
Teetes GL (1996). Plant resistance to insects: a fundamental component of IPM. Radcliffe's IPM world textbook'.(Eds EB Radcliffe, WD Hutchison, RE Cancelado)(University of Minnesota: St Paul)].
|
|
|
|
|
Thottappilly G, Rossel HW (1987). Seed transmission of cowpea (yellow) mosaic virus unlikely in cowpea. (IITA) Trop. Grain Leg. 34:1987.
|
|
|
|
|
Umaharan P, Ariyanayam RP, Haque SQ (1997). Resistance to cowpea severe mosaic virus, determined three dosage dependent genes in Vigna unguiculata L. Walp. Euphytica 95:49-55.
Crossref
|
|
|
|
|
Uyemoto JK, Claflin LE, Wilson DL, Raney RJ (1981). Maize chlorotic mottle and maize dwarf mosaic viruses; effect of single and double inoculations on symptomatology and yield. Plant Dis. 65(1):39-41.
Crossref
|
|
|
|
|
Van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB (2000). Virus Taxonomy: Seventh Report of the International Committee on Taxonomy of Viruses. San Diego: Academic Press.
|
|
|
|
|
Zettler FW, Evans IR (1972). Blackeye cowpea mosaic virus in Florida: host range and incidence in certified cowpea seed. Paper presented at the Proceedings of the Florida State Horticultural Society.
|
|