Full Length Research Paper
ABSTRACT
Yield is a quantitative trait known to be influenced by changes in the environment in which the crop is grown, suggesting the need to evaluate soybean lines in different growing regions to assess their adaptability and stability. The objective of this study was to evaluate the performance, yield stability, and genotype × environment interaction (G×E) in soybean cultivars, and the informativeness of six environments in Southern Africa. A group of 62 soybean lines was grown and tested in six locations in Malawi and Zambia during the growing seasons in 2018 and 2019 using a 6 x 5 and 5 x 5 alpha lattice design. Genotype plus genotype × environment (GGE) biplots were used to reduce data complexity and analyze genotypes performance and stability; environments discriminativeness and representativeness; and traits associated with yield. The GGE analyses showed a significant effect of genotypes, environments, and G×E. Genotypes with local and wide adaptation and environments with desirable characteristics for testing cultivars were identified. A number of agronomic traits were identified to be positively and negatively associated with soybean yield. These results reinforce the utility of GGE analysis for ranking cultivars and environments in soybean breeding programs in Southern Africa.
Key words: GGE, yield, adaptation, stability, heritability, flowering.
Abbreviation: AMMI, Additive Main effect and Multiplicative Interaction; GGE, Genotype plus genotype by environment; G X E, genotype by environment interactionINTRODUCTION
Soybean (Glycine max (L.) Merr.) is an important protein and oilseed crop throughout the world. Its high protein content (40 - 42%) makes it suitable and desirable in human diet (Patil et al., 2018). Soybean seeds have an oil content of approximately 18 to 22% (Patil et al., 2018) making it a major source of both edible and inedible oil products. Its high-quality oil is used in food product manufacturing as well as in the production of inedible products such as caulks, mastics, plastics, and resins. Additionally, when integrated with a rotational system with cereals, it enhances the level of nitrogen in the soil (Kumudini, 2010) playing an important role in soil fertility.
Soybean is grown in most parts of the world, with Brazil being the leading producer, followed by the USA and then Argentina. Africa produces 12,537 tons (about 2.2% of the global production) with South Africa being the leading soybean producer followed by Nigeria and then Zambia (FAOSTAT, 2019). Soybean production has gradually been increasing in Zambia making it the leading exporter to southern African countries such as Zimbabwe and Botswana (Abate et al., 2012). However, further growth of Zambia’s soybean export market within the southern African region is constrained by its low yields. One of the contributing factors to low yields is the limited availability of varieties that are adapted to local and regional climatic conditions.
Soybean yield is controlled by multiple genes and it is affected by the interactions between the genotype and the environment, in previous studies soybean seed yield has been found to be associated with up to 17 QTL (Han et al., 2012). Varietal stability is a key factor affecting cultivar’s response in varying environments. Stability analysis aims at examining the performance of a genotype relative to other genotypes in different environments (Bernardo, 2020). Breeders may require a variety that is stable across different production environments; however, there could be a genotype that may perform well in a specific environment but poorly in other environments. This lack of varietal stability may be challenging for breeding programs driven by the development of soybean varieties adapted to multiple environments in multiple countries such as the soybean breeding program of the International Institute of Tropical Agriculture (IITA). Therefore, it is important to determine the stability of a given genotype in multiple production environments to inform variety release decisions.
To fully express their genetic potential, cultivars require ideal environmental conditions; hence, their performance will vary depending on the production environment (Reynolds et al., 2001). This relative change in performance of cultivars across environments is termed genotype by environment interaction (G×E). G×E reduces the association between phenotypic and genotypic values, which may cause selections from one environment to perform poorly in another. An understanding of G×E in multi-environment trials (MET) is important in identifying locations that are efficient in distinguishing tested genotypes and in providing information on the most representative and descriptive environment.
As G×E complicates the identification of ideal environments, and superior and stable genotypes, multiple data reduction tools such as cluster analysis, joint plots and biplots have been used by breeders (Yan and Tinker, 2006). Biplots allow the identification of associations between genotypes, environments, and their interaction. Biplots include the Additive Main effect and Multiplicative Interaction (AMMI) and Genotype plus genotype by environment (GGE) analyses. GGE biplots are powerful tools that combine principal component analysis (PCA) and graphical explanation of G×E to identify patterns associated with genotypes and G×E in the evaluation of cultivars in MET (Hoyos-Villegas et al., 2016. GGE allows a better understanding of G×E, and in turn facilitates the identification of representative and discriminative environments (Yan and Tinker, 2006). In contrast, AMMI analysis explain less of the GGE variation compared to GGE biplots, and does not allow an accurate visualization of genotypes in a given environment since the inner-product property is not considered (Hoyos-Villegas et al., 2016; Yan et al., 2007). The objective of this study was to assess the performance, yield stability, G×E in soybean cultivars, and the informativeness of the tested environments in multiple locations in Southern Africa.
MATERIALS AND METHODS
Genotypes and evaluation environments
Fifty-five (F7) elite soybean lines developed by the International Institute of Tropical Agriculture (IITA) along with seven commercial checks (released varieties) from seed companies were used to generate the data used in this study. The genotypes being evaluated have an early to medium maturity group with determinate and indeterminate growth habit (Table 1). The multi-environment trials (MET) were carried out at six different locations in Zambia and Malawi. In 2018 season, the locations were Chipata, Seedco-Zambia, International Institute of Tropical Agriculture Southern African Region Administration (IITA-SARAH), and Chitedze (Malawi). In 2019 season, the locations were Bvumbwe (Malawi), Kabwe (Zambia), IITA-SARAH, and Chitedze (Malawi). According to their characteristics, locations were grouped into four environments (Table 2).
Experiment design and data collection
The soybean genotypes were planted in a 6 × 5 and 5 × 5 alpha lattice design with three replications. Each genotype occupied a plot comprising four rows of 4 m long each, 0.5 m between rows, and 0.05 m between plants. Basal dressing fertilizer (25 kg N/ha, 30 kg K2O/ha, 60 kg P2O5/ha) was applied at planting, and Metolachlor and Imazethapyr were applied as pre-emergence herbicides for control of weeds. Quizalofop-p-ethyl and Fomesafen were applied as post-emergence control of weeds and hand weeding control was also done. Data collected involved grain yield (GY, kg ha-1), 100 seed weight (SWT100), plant height (PLHT), days to maturity (DM), days to flowering (DFFL), lodging, and shattering. Lodging was recorded for each plot based on a descriptive scale (Woods and Swearingin, 1977) where 1 = almost all plants erect, 2 = either all plants leaning slightly or a few plants down, 3 = either all plants leaning moderately (<45° angle) or 25-50% of plants down, 4= either all plants leaning considerably (>45° angle) or 50-80% of plants down, and 5 = all plants down. For shattering the scale proposed by Krisnawati and Muchlish Adie (2017) was used, where 1 = No pod-shattering (Very Resistant); 2 ≤ 25% pod-shattering (Resistant); 3 = 25-50% pod-shattering (Moderately Resistant); 4 = 51-75% pod-shattering (Highly Susceptible); 5 ≥75% pod-shattering (Very Highly Susceptible).
Statistical analysis
Genotype plus Genotype × Environment biplots and stability analysis
Datasets from Chipata, IITA-SARAH, Chitedze, and Seedco-Zambia in 2018 were combined with datasets from Chitedze, IITA-SARAH, Bvumbwe, and Kabwe in 2019, respectively, to establish four environments (Table 1). Normality was also checked in data using residuals.
Multi-environment data were analyzed to visually identify GE interactions. The package metan in R (Olivoto and Lúcio, 2020) was used to calculate ANOVA-based stability statistics, variance components, and broad-sense heritability (H2) in mixed-effect models, and best linear unbiased prediction (BLUP) values for GY, SWT100, PLHT, DM, DFFL, lodging, and shattering, using the model:
where µ is the grand mean, 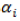 is the random effect of the ith line (genotype),
is the random effect of the ith line (genotype), 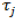 is the random effect of the jth environment,
is the random effect of the jth environment, 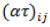 is the interaction of genotype i and environment
is the interaction of genotype i and environment 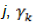 is the random effect of the kth block within the j environment, and
is the random effect of the kth block within the j environment, and 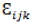 is the random error. BLUP-adjusted means were obtained by resolving the random effects for each line (Olivoto and Lúcio, 2020).
is the random error. BLUP-adjusted means were obtained by resolving the random effects for each line (Olivoto and Lúcio, 2020).
Different biplots were created using the environment-centered model 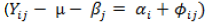 where the E main effect
where the E main effect 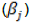 is removed and the biplot only considers G
is removed and the biplot only considers G 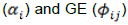 as the relevant sources of variation. The biplot analyses were carried out using the package GGE Biplot GUI in R (Frutos et al., 2014). In the biplots, no scaling was used, except for the genotype by trait biplot where data were scaled with the standard deviation (SD). When evaluating test environments or traits, data were represented using column-preserving, singular-value; whereas, forthe comparison of G and GE, biplots were drawn using row-preserving, singular value partitioning (Yan and Tinker, 2006).
as the relevant sources of variation. The biplot analyses were carried out using the package GGE Biplot GUI in R (Frutos et al., 2014). In the biplots, no scaling was used, except for the genotype by trait biplot where data were scaled with the standard deviation (SD). When evaluating test environments or traits, data were represented using column-preserving, singular-value; whereas, forthe comparison of G and GE, biplots were drawn using row-preserving, singular value partitioning (Yan and Tinker, 2006).
RESULTS AND DISCUSSION
Average agronomic performance, analysis of variance and heritability
Grain yield in E1 ranged from 1.89 t/ha (TGx2001-8FM) to 2.86 t/ha (TGx2014-9FM) with an average of 2.33 t/ha. In E2, grain yield ranged from 2.20 t/ha (TGx2001-19DM) to 2.99 t/ha (SC SQUIRE) with an average of 2.56 t/ha. E3 had a grain yield ranging from 1.71 t/ha (TGx2001_9DM) to 2.69 t/ha (TGx2014_24FM) with an average of 2.24 t/ha. In E4 grain yield ranged from 2.92 t/ha (TGx2001_8DM) to 3.58 t/ha (TGx2001_2DM) with an average performance of 3.21 t/ha. The agronomic performance of traits across environments showed that grain yield ranged from 2.27 t/ha (TGx2001-8FM) to 2.88 t/ha (TGx2014_24FM) with an average of 2.58 t/ha. Similar findings were reported, in which soybean yield ranged from 1.77 (SC Semeki) to 2.43 t/ha (Lukanga) with an average of 1.95 t/ha (Manda and Maata, 2020). SWT100 ranged from 13.10 g (Dina) to 17.96 g (TGx2001_14DM) with an average of 14.88 g, PLHT ranged from 67.49 cm (TGx2002_14DM) to 85.19 cm (TGx2014_23FM) with an average of 75.35 cm, DM oscillate between 103 days (TGx2014_31FM) and 118 days (Dina) with an average of 111 days; these results indicate that most of our genotypes have a medium maturity period, following the scale developed by Kawuki et al. (2003), in which early maturing varieties takes 85 to 99 days, medium maturing varieties take 100 to 111 days to maturity and late maturing varieties take 112 to 121 days to maturity, DFFL varied from 45 days (TGx2014_31FM) to 56 days (TGx1987_62F) with an average of 51 days, lodging score ranged from 1.82 (TGx2014_24FM) to 2.41 (TGx2002_4DM) with an average of 2.09, and shattering score was between 1.17 (Dina) and 2.38 (TGx2002_6FM) with an average of 1.68.
In the analysis of variance, all the sources of variation had a significant effect (p<0.001) on GY, DM, DFFL, and shattering. The environment did not have a significant effect on PLHT or lodging. Lodging showed the highest variation (CV = 58.7%) followed by shattering (CV = 38.4%). In contrast, DM and DFFL had the lowest CV with 4.8 and 8.5%, respectively (Table 3).
Table 4 shows the proportion of variance explained by the Environment, Genotype, and G×E interaction, and the broad-sense heritability (H2) for the traits evaluated. Overall, the environment explained a higher proportion of the variance, accounting for 4.56 to 50.03%, followed by G×E (5.03-18.06%), and the genotype (0.54-15.46%). For yield, the environment explained 14.36% of the variability, genotype 1.02%, and the interaction 5.06%. The magnitude of the contribution of the E and G×E are attributed to contrasting conditions between testing environments (Ahakpaz et al., 2021). In the present study, environments in the two countries vary in terms of soil type, precipitation, temperature, and humidity (Table 2), differences that explain the observed effects. Previous studies have also reported a higher contribution of the environment than G×E on soybean yield (Gurmu et al., 2017; Rakshit et al., 2012; Temesgen et al., 2015; Vaezi et al., 2017) and other traits (Li et al., 2020). Nevertheless, higher G×E contributions (Atnaf et al., 2013; Bhartiya et al., 2018; Mwiinga et al., 2020) or a higher genotype effect (Li et al., 2020) has also been reported.
The H2 can be divided into high (H2 greater than 50%), medium (H2 greater than 20% and less than 50%), and low (H2 less than 20%) (Sulistyo et al., 2018). According to this, GY, SWT100, and lodging showed a medium heritability (37, 44, and 21% respectively), and PLHT, DM, DFFL, and shattering showed a high H2 (50-76%; Table 4). For yield-related variables, the H2 was lower compared to those reported by Li et al. (2020) for soybean MET in China. However, our H2 values for yield and other variables are consistent with previous reports that showed PLHT, DM, and DFFL having a higher heritability compared to yield in soybean (Bianchi et al., 2020; Li et al., 2020).
GGE analysis
Although the effect of the environment explained a large percentage of the total variance (Table 4), significant contributions of G×E to total variation for all the variables evaluated were also identified (Table 3). When selecting stable genotypes and evaluating tester environments, the effect of the genotype and G×E are the main factors to be examined simultaneously (Yan and Kang, 2003; Yan et al., 2007). Here, the GGE analysis allowed to graphically evaluate the environments and genotypes.
Environment evaluation
The discriminativeness vs representativeness is a tester-centered (G + GE) and column-preserving biplot that assesses environments. In this biplot, it is possible to identify environments that are similar and could be grouped into mega-environments. Similarly, the biplot provides information about which of the environments is more representative within the trials and which one has the best discriminating ability. Relationships among the four environments can be visualized by analyzing their vectors and the angles formed among them. Acute angles indicate positive correlations and obtuse angles negative correlations. All the environments showed a positive correlation, where the angles formed among the environment vectors allowed to identify two major groups, one with E2, E3, and E4 and another with E1 (Figure 1). However, E4 is close to being in the middle between E2-E3 and E1. Information on mega-environments allows a better experimental design for the evaluation of multiple genotypes and to determine when locations can be predictable for a given environment (Gauch et al., 2008). As reported here, GGE and other biplot analysis such as AMMI have shown to be efficient to identify mega environments (Ahakpaz et al., 2021; Hoyos-Villegas et al., 2016; Li et al., 2020). The mega-environments identified here allow the use of repeatable G×E in soybean breeding in Southern Africa, and contribute to the crop production by releasing adapted cultivars according to mega-environments (Yan, 2019). These results also demonstrate the utility of GGE biplots to analyze multiple locations and their relevance in the routine MET conducted in breeding programs aimed to release cultivars with wide adaptation.
The concentric circles on the biplot are proportional to the standard deviation and help to visualize the length of the vectors. Environments with longer vectors have higher discriminating ability. Thus, E1 and E3 were the most discriminating environments followed by E2 and E4, which had shorter vectors and may provide less information on the evaluated genotypes (Figure 1). In Figure 1, the average environment, indicated with an arrowhead and a circle, and an Average-Environment Axis (AEA) have also been included. The AEA is a line that passes through the average environment and the biplot origin and helps to categorize the environments based on their representativeness. A test environment with the smallest angle with the AEA is more representative. Therefore, E4 is the most representative environment, followed by E2, E3, and E1 (Figure 1). Test environments that are both representative and discriminating are useful to select genotypes that have general adaptation. In contrast, discriminating but non-representative environments are valuable to select genotypes with specific adaptation if the environment can be divided into mega-environments or eliminate unstable genotypes if the target environment is a single mega-environment (Yan and Tinker, 2006). Here, the testing environments composed of single environments as described in materials and methods, hence E1 and E3, which were the most discriminating, but were not representative, would be useful environments to identify locally adapted genotypes and potential parents.
The ideal environment biplot ranks the tester environments based on discriminating ability and representativeness considering an “ideal environment”. An ideal environment is that with the highest discriminativeness and representativeness. Figure 2 shows the “ideal environment” with the arrowed line in the center of the concentric circles and it is located on the AEA with a distance from the biplot origin equal to the longest vector. The E3 was the closest environment to an ideal, followed by E1, E2, and E4. The results are consistent with previous reports in multiple crop species where GGE biplots have been successfully applied to characterize environments that allow a better selection of genotypes and the optimization of resources in breeding programs (Baraki et al., 2020; Luo et al., 2015; Mwiinga et al., 2020).
Genotype evaluation
The performance of genotypes in each environment can be analyzed by comparing the angles formed among the vectors of genotypes and environments. Acute angles indicate that a genotype performs better than average in a specific environment, obtuse angles suggest that the performance is lower than average, and if a right angle is formed between a genotype and an environment, the performance of the genotype is close to the average in that environment (Hoyos-Villegas et al., 2016). TGx2002_35FM, TGx2014_24FM, and TGx2001_11DM yielded better than average in E2, E3, and E4, while TGx2014_9FM, Squire, TGx2014_33FM, and TGx2001_16DM are better adapted to E1 (Figure 1).
The origin of the biplot is referred to as a “virtual” genotype which has a performance close to the average in all environments (Hoyos-Villegas et al., 2016). Among the evaluated genotypes, TGx2001_13FM and TGx2014_16FM were near the biplot origin, which means that these genotypes have a lower contribution to genotype or GE interactions (Figure 1). Genotypes with vectors distal from the biplot origin would have higher contributions to genotype or GE interactions with a better or worse response across all environments depending on the direction of the vectors. TGx2014_24FM, TGx2002_17DM, and TGx2014_9FM had the longest vectors in the same direction as the environments, indicating that these genotypes had the best performance across all environments. In contrast, TGx2001_19DM, TGx2001_8FM, Dina, and TGx2001_14DM had the longest vectors in the opposite direction of the environments and thus, the lowest performance across E1 – E4. Our results are consistent with those reported by Mwiinga et al. (2020)who evaluated some of the lines used in the present study, and found that TGx2014_24FM and TGx2002_17DM showed a positive performance in different environments.
By examining the angle formed between the AEA and vector of the genotype, it is possible to observe if the response of a genotype is mainly due to GE effects. Genotypes such as Safari, TGx2002_5FM, TGx2014_5GM, and TGx2002_14DM showed right angles with the AEA, indicating that their response can be mainly attributed to GE interactions (Figure 1). Regarding the elite genotypes, Dina and Lukanga had lower than average performance in all environments, whereas Kafue had a better performance in E1 and Safari in E2 – E4.
The mean performance and stability biplot is appropriate for genotype evaluation. In this biplot, it is possible to identify the most stable genotype and the one with the highest mean performance. Figure 3 shows the Average-Environment Coordination (AEC) view of the genotype-metric preserving biplot. On the AEC, genotypes that are closer or above the AEC abscissa (single-arrowed line) in the same direction of environments, are expected to have the highest mean performance. In contrast, genotypes located farther from the AEC abscissa arrowhead would have the lowest performance. The genotypes TGx2014_24FM, TGx2014_9FM, TGx2001_11DM, and TGx2014_4FM had the highest mean performance, and TGx2001_8FM, TGx2001_19DM, and TGx2001_14DM the lowest (Figure 3).
The AEC ordinate on axis y (double-arrowed line) points to poorer stability in either direction and genotypes near the AEC ordinate have a mean yield similar to the grand mean. Longer vectors correspond to genotypes with poorer stability and genotypes with short vectors are expected to be more stable. TGx2002_17DM, TGx2002_3FM, and TGx2002_23DM were found to be the genotypes with the greatest variability, compared to TGx2002_4DM, TGx2002_8FM, TGx2001_11DM, and TGx2014_24FM that were highly stable. Similar results have been reported by Mwiinga et al. (2020)who found, for instance, that TGx2002_17DM, had the lowest yields in some environments and was the winner in some of the environments evaluated. In this way, our results further support previous findings and highlighting how variable genotypes can be successfully identified by the analyses performed here.
Six genotypes, including TGx2002_5FM, Safari, TGx2001_13FM, Kafue, TGx2002_14DM, and TGx2001_24FM performed at the grand mean (Figure 3). It is worth noting that the two first principal components explained 71.4% of the variation, which may not reflect the actual stability for some of the genotypes (Hoyos-Villegas et al., 2016). However, the variability captured in our analysis is slightly higher than previously reported in a similar experiments using AMMI biplots (Mwiinga et al., 2020).
Based on the performance and the stability, genotypes can be ranked relative to an “ideal” genotype, which is that with the highest performance and the lowest variability. Among the genotypes, TGx2014_24FM was the closest to an ideal genotype, followed by TGx2001_11DM (Figure 4). Therefore, TGx2014_24FM and TGx2001_11DM could be considered for release in multiple environments. Interestingly, the elite lines (Dina, Kafue, Lukanga, and Safari) showed yields lower or close to the grand mean and were considerably unstable, being far from the ideal environment (Figures 3 and 4). In the characterization of genotypes, GGE biplots have been shown to be powerful tools to assess the stability and adaptation, and to demonstrate how these parameters can be increased by the incorporation of other sources of information in breeding programs (Merrick et al., 2020), highlighting the applicability of the method and our results.
The which-won-where pattern shown by a GGE biplot represents concepts such as crossover GE, mega-environment differentiation, and specific adaptation (Hoyos-Villegas et al., 2019; Weikai Yan and Tinker, 2006). The which-won-where biplot is constructed by drawing a polygon on genotypes that are furthest from the origin. The polygon will contain all the genotypes, and perpendicular lines to the sides allow defining sectors where the performance of the genotypes within environments can be analyzed. Genotypes within the same sector of an environment are expected to have a better response in that environment. A total of six sectors were identified, and the environments fell into two sectors (Figure 5). E2 - E4 were located in the same sector, where TGx2014_24FM had the best performance (genotype located at one of the vertices of the polygon), followed by TGx2001_11DM. In comparison, in the E1, the genotype with the best response was TGx2014_9FM, followed by TGx2001_24FM. Therefore, these genotypes are recommended to be released in E2 - E4 or in E1, respectively. Using GGE evaluation, TGx2014_24FM among other genotypes with a similar response, was identified as a genotype that showed a wider adaptation to more than one environment. This is consistent with Mwiinga et al. (2020)who also were able to identify genotypes with wide adaptation.
Genotype-by-trait analysis
Yield is a major breeding objective, however, breeding for yield tends to be challenging because of its genetic complexity, low heritability, and the effect of G×E interactions (Bianchi et al., 2020). Besides dissecting yield by decomposing the variance attributable to E, G and G×E, and the analysis of the G×E through GGE analysis, the identification of correlated variables can contribute to improving single yield components, and then the overall yield performance (Shahriari et al., 2018). Furthermore, strongly correlated variables with high heritability could help to indirectly select for yield and circumvent challenges of direct selection.
Multiple traits averaged across all environments can be correlated with genotypes using genotype × trait biplots. These biplots allow to visually recognize relationships between variables, genotypes, and their interactions; being possible to identify traits that are positively or negatively associated, traits that could be redundant or that can be used as indirect indicators of other measurements (Yan and Tinker, 2006). Following the same approach in angle formation used for environment and genotype evaluation, PLHT, DM, and DFFL were positively correlated to each other and were negatively correlated to shattering (Figure 6). Regarding lodging, DFFL, PLHT, and shattering were positively correlated, but DM was independent. Yield variables (GY and SW100) were grouped in the biplot and showed a strong negative relation with lodging; and a slight negative correlation with shattering, PLHT, DM and DFFL. Despite GY and SWT100 had medium H2, correlated traits such as PLHT, DM, and DFFL had high H2, reinforcing that they could be used for indirect selection for yield. Our results complement previous studies on soybean breeding programs in Southern Africa (Mwiinga et al., 2020), where the correlation between yield and other traits was not evaluated. Interestingly, the negative correlation found between yield and PLHT differs from previous results in soybean where selection for plants with higher plant height improved overall yield (Li et al., 2020), which could be attributed to the adaptation of breeding lines to the environmental conditions in Southern Africa. In the cultivar development, further MET analyses aimed to address these correlations are necessary to incorporate them as indirect selection strategies. Considering associated variables can simplify breeding programs and yield improvement as redundant measurements, variables that may be easier to assess, and indirect sources of information can be identified, which makes genotype × trait analysis a valuable decision-making tool for soybean breeding (Hoyos-Villegas et al., 2016; Li et al., 2020).
Regarding genotypes, TGx2014_24FM, TGx2002_14DM, and TGx2014_9FM had the highest GY and SW100, and the lowest lodging, these could be used as potential parents for improving yield and reduce lodging score. On the other hand, TGx2001_11DM and Squire, which had considerable yield performance, had a lower shattering score, which did not have an apparent effect on TGx2014_24FM and TGx2002_14DM. Among the checking lines, Safari and Lukanga showed a higher GY and SW100. These findings are consistent with previous studies where, as a checking line, Lukanga had shown to have a higher yield response to different environmental factors such as population density. Lukanga and Kafue had a higher shattering score and Dina had a higher PLHT, DM, and DFFL, accompanied by a lower GY, SW100, and shattering.
CONCLUSIONS
In the present study, GGE analysis revealed that the yields of the 62 soybean genotypes tested were significantly affected by genotype, environment, and genotype by environment. E1 (Chipata and Chitedze) and E3 (Bvumbwe) were identified as the most discriminating environments, and E4 (Seedco.Zambia and Kabwe) as a highly representative environment. TGx2014_24FM was identified as the most ideal genotype, which also had the highest yield across environments (2.88 t/ha), followed by TGx2001_11DM (2.86 t/ha). Through genotype × trait analysis, it was identified that yield was positively associated with plant height, days to flowering, and days to maturity, negatively associated with lodging, and independent of shattering. Our results support the used GGE analysis as an analysis tool to dissect complex traits largely affected by G×E such as yield by decomposing its variance and by identifying related variables.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
ACKNOWLEDGMENTS
The authors would like to thank the International Institute of Tropical Agriculture IITA-Zambia through the soybean breeding program for the financial support and providing experimental lines. The opinions expressed in this publication are those of the authors and do not necessarily reflect the views of the IITA.
REFERENCES
|
Abate T, Alene AD, Bergvinson D, Shiferaw B, Silim S, Orr A, Asfaw S (2012). Tropical grain legumes in Africa and South Asia: knowledge and opportunities. Nairobi, Kenya: International Crops Research Institute for the Semi-Arid Tropics. 112 pp. |
|
|
Ahakpaz F, Abdi H, Neyestani E, Hesami A, Mohammadi B, Mahmoudi KN, Abedi-Asl G, Noshabadi MRJ, Ahakpaz F, Alipour H (2021). Genotype-by-environment interaction analysis for grain yield of barley genotypes under dryland conditions and the role of monthly rainfall. Agricultural Water Management 245(12):2020. |
|
|
Atnaf M, Kidane S, Abadi S, Fisha Z (2013). GGE biplots to analyze soybean multi-environment yield trial data in north Western Ethiopia. Journal of Plant Breeding and Crop Science 5(12):245-254. |
|
|
Baraki F, Gebregergis Z, Belay Y, Berhe M, Teame G, Hassen M, Gebremedhin Z, Abadi A, Negash W, Atsbeha A, Araya G (2020). Multivariate analysis for yield and yield-related traits of sesame (Sesamum indicum L.) genotypes. Heliyon 6(10):e05295. |
|
|
Bernardo R (2020). Reinventing quantitative genetics for plant breeding: something old, something new, something borrowed, something BLUE. Heredity 125:6. |
|
|
Bhartiya A, Aditya JP, Kumari V, Kishore N, Purwar JP, Agrawal A, Kant L (2018). Stability analysis of soybean [Glycine max (L.) Merrill] genotypes under multi-environments rainfed condition of North Western Himalayan hills. Indian Society of Genetics and Plant Breeding 78(3):342-347. |
|
|
Bianchi MC, Bruzi AT, Soares IO, Ribeiro FDO, Gesteira GDS (2020). Heritability and the genotype × environment interaction in soybean. Agrosystems, Geosciences and Environment 3(1):1-10. |
|
|
FAOSTAT (2019). Agriculture Organization of the United Nations Statistics Division (2018). |
|
|
Frutos E, Galindo MP, Leiva V (2014). An interactive biplot implementation in R for modeling genotype-by-environment interaction. Stochastic Environmental Research and Risk Assessment 28(7):1629-1641. |
|
|
Gauch HG, Piepho HP, Annicchiarico P (2008). Statistical analysis of yield trials by AMMI and GGE: Further considerations. Crop Science 48(3):866-889. |
|
|
Gurmu F, Hussein S, Laing M (2017). Genotype-by-environment interaction and stability of sweetpotato genotypes for root dry matter, β-carotene and fresh root yield. Open Agriculture 2(1):473-485. |
|
|
Hoyos-Villegas V, O'Connor JR, Heslop AD, Hilditch A, Jahufer MZZ, Barrett BA (2019). Rate of genetic gain for persistence to grazing and dry matter yield in white clover across 90 years of cultivar development. Crop Science 59(2):537-552. |
|
|
Hoyos-Villegas V, Wright EM, Kelly JD (2016). GGE biplot analysis of yield associations with root traits in a mesoamerican bean diversity panel. Crop Science 56(3):1081-1094. |
|
|
Kawuki RS, Adipala E, Tukamuhabwa P (2003). Yield Loss Associated with Soya Bean Rust (Phakopsora pachyrhizi Syd.) in Uganda. Journal of Phytopathology 151(1):7-12. |
|
|
Krisnawati A, Muchlish Adie M (2017). Characterization and performance of agronomic characters of soybean genotypes resistant to pod shattering. Biodiversitas 18(3):1158-1164. |
|
|
Kumudini S (2010). Soybean growth and development. The Soybean: Botany, Production and Uses pp. 48-73. |
|
|
Li M, Liu Y, Wang C, Yang X, Li D, Zhang X, Xu C, Zhang Y, Li W, Zhao L (2020). Identification of Traits Contributing to High and Stable Yields in Different Soybean Varieties Across Three Chinese Latitudes. Frontiers in Plant Science 10(1):1-14. |
|
|
Luo J, Pan YB, Que Y, Zhang H, Grisham MP, Xu L (2015). Biplot evaluation of test environments and identification of mega-environment for sugarcane cultivars in China. Scientific Reports 5(9):1-11. |
|
|
Manda N, Maata M (2020). Responses of soybeans (Glycine max (L.) Merrill) associated with variable plant density stress applied at different phenological stages: Plasticity or elasticity? African Journal of Biotechnology 19(6):307-319. |
|
|
Merrick LF, Lyon SR, Balow KA, Murphy KM, Jones SS, Carter AH (2020). Utilization of evolutionary plant breeding increases stability and adaptation of winter wheat across diverse precipitation zones. Sustainability (Switzerland) 12(22):1-23. |
|
|
Mwiinga B, Sibiya J, Kondwakwenda A, Musvosvi C, Chigeza G (2020). Genotype x environment interaction analysis of soybean (Glycine max (L.) Merrill) grain yield across production environments in Southern Africa. Field Crops Research 256(5):107922. |
|
|
Olivoto T, Lúcio ADC (2020). metan: An R package for multi-environment trial analysis. Methods in Ecology and Evolution 11(6):783-789. |
|
|
Patil G, Vuong TD, Kale S, Valliyodan B, Deshmukh R, Zhu C, Wu X, Bai Y, Yungbluth D, Lu Y, Kumpatla S, Shannon JG, Varshney RK, Nguyen HT (2018). Dissecting genomic hotspots underlying seed protein, oil, and sucrose content in an interspecific mapping population of soybean using high-density linkage mapping. Plant Biotechnology Journal 16(11):1939-1953. |
|
|
Rakshit S, Ganapathy KN, Gomashe SS, Rathore A, Ghorade RB, Kumar MVN, Ganesmurthy K, Jain SK, Kamtar MY, Sachan JS, Ambekar SS, Ranwa BR, Kanawade DG, Balusamy M, Kadam D, Sarkar A, Tonapi VA, Patil JV (2012). GGE biplot analysis to evaluate genotype, environment and their interactions in sorghum multi-location data. Euphytica 185(3):465-479. |
|
|
Reynolds MP, Calderini DF, Condon AG, Rajaram S (2001). Physiological basis of yield gains in wheat associated with the Lr19 translocation from Agropyron elongatum. Euphytica 119(1):137-141. |
|
|
Shahriari Z, Heidari B, Dadkhodaie A (2018). Dissection of genotype × environment interactions for mucilage and seed yield in Plantago species: Application of AMMI and GGE biplot analyses. In PLoS ONE 13(5):e0196095. |
|
|
Sulistyo A, Purwantoro, Sari KP (2018). Correlation, path analysis and heritability estimation for agronomic traits contribute to yield on soybean. IOP Conference Series: Earth and Environmental Science 102(1). |
|
|
Temesgen T, Keneni G, Sefera T, Jars M (2015). Yield stability and relationships among stability parameters in faba bean (Vicia faba L.) genotypes. Crop Journal 3(3):258-268. |
|
|
Vaezi B, Pour-Aboughadareh A, Mohammadi R, Armion M, Mehraban A, Hossein-Pour T, Dorii M (2017). GGE Biplot and AMMI analysis of barley yield performance in Iran. Cereal Research Communications 45(3):500-511. |
|
|
Woods SJ, Swearingin ML (1977). Influence of Simulated Early Lodging upon Soybean Seed Yield and its Components. Agronomy Journal 69(2):239-242. |
|
|
Yan W (2019). LG biplot: a graphical method for mega-environment investigation using existing crop variety trial data. Scientific Reports 9(1):1-8. |
|
|
Yan W, Kang MS (2003). GGE Biplot Analysis: A Graphical Tool for Breeders, Geneticists, and Agronomists. CRC Press. |
|
|
Yan W, Kang MS, Ma B, Woods S, Cornelius PL (2007). GGE biplot vs. AMMI analysis of genotype-by-environment data. Crop Science 47(2):643-655. |
|
|
Yan W, Tinker NA (2006). Biplot analysis of multi-environment trial data: Principles and applications. Canadian Journal of Plant Science 86(3):623-645. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0