ABSTRACT
A cross-sectional study was conducted between October 2014 and May 2015, to determine the sero-prevalence, assess the presence of active cases through isolation and identify possible risk factors for the Middle East Respiratory Syndrome Corona Virus (MERS-CoV) in camels, in selected areas of Oromia and Afar regional states of Ethiopia. A total of 569 dromedary camel sera were collected and screened with two serological tests: Pseudo particle neutralization for screening and Micro-neutralization test for confirmation. The overall prevalence of MERS-CoV in the study area was 93% (n=529). Higher prevalence (93.9%) was recorded in female dromedary camels compared to males (91.3%) but the difference was not significant (Chi-Square=1.323 and P=0.250). Age wise prevalence was higher in adult camels (94.6%) than young ones (90.8%), however the difference was statistically insignificant (Chi-Square=3.068 and P=0.080). Similarly, higher prevalence was recorded in larger herds (93.4%) than small herds but the difference observed was not statistically significant (Chi-Square=0.220 and p=0.639). Also, no significant (Chi-Square 0.525 and P=0.469) difference was observed in prevalence between the two regions (Oromia=93.3% and Afar=93.3%). However significant difference was observed in the lower administrations of the regions (Zones, District and Kebeles) in which the highest prevalence recorded in East Shoa, Fentale district of Adis Ketema kebele (99.3%). From total of 100 swabs collected, MERS-CoV was detected in seven (1 Fentale, 4 Amibara and 2 Dubti) districts by Real-time reverse transcription polymerase chain reaction (RT-PCR). Generally, this study showed the existence of high seroprevalence of MERS-CoV among Ethiopian dromedary camels, which was also confirmed by RT-PCR. Therefore further study is required to determine its significance from both animal and public health perspectives and further research should focus on identifying similarity between MERS-CoV viral isolates in neighboring countries and clinical isolates from the Middle East and elsewhere.
Key words: Dromedary camel, MERS-CoV, serum, swab, Real-time reverse transcription polymerase chain reaction (RT-PCR), Ethiopia.
More than 60% of the world population of one-humped dromedary camels (Camelus dromedarius) lives in the Greater Horn of Africa. Their role as one of the most important livestock species for nutrition in arid and semi-arid areas of Eastern Africa is likely to increase since the predicted climate change is in favour of this drought resilient species. Camel milk, for domestic consumption or sale, is often the most important product for female and male pastoralists alike (Marshall et al., 2014). In 2012, a novel coronavirus called Middle East Respiratory Syndrome Coronavirus (MERS-CoV) emerged on the Arabian Peninsula. Human cases have been reported from 25 countries, with the most recent outbreak in the Republic of Korea (which itself originated in the Middle East). MERS-CoV has caused at least 1,200 laboratory confirmed cases of severe respiratory infection, including more than 400 deaths. Several studies have demonstrated that dromedary camels can act as a source of human MERS-CoV infection (Reusken et al., 2015).
In June 2012, a novel beta corona virus, associated with severe respiratory disease in humans, emerged in the Middle East (Zaki et al., 2012), which is closely related to beta corona viruses, circulating in bats (Anthony et al., 2013). The first isolate of Middle East respiratory corona virus (MERS-CoV), HCoV-EMC/2012, was obtained from a patient with a fatal pneumonia and acute renal failure. To date, 107 additional human cases have been identified, of which 49 were fatal (WHO, 2013). Aside from cases in Saudi Arabia, Qatar, Jordan, and the United Arab Emirates, imported cases have been identified in the United Kingdom, Germany, France, Tunisia, and Italy.
Although no information is available on the source or route of primary transmission of MERS-CoV, human-to-human transmission has been recorded (Assiri et al., 2013). The severity of disease distinguishes MERS-CoV from other corona viruses, circulating in the human population HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV HKU1, which are generally associated with upper respiratory tract infections. Instead, MERS-CoV appears to be more similar to the severe respiratory disease, caused by severe acute respiratory syndrome (SARS)-CoV. In vitro studies have shown that MERS-CoV replicates efficiently in non-ciliated cells in the primary human airway epithelium (Kindler et al., 2013), and in ex vivo human lung cultures, MERS-CoV replicated in bronchial, bronchiolar, and alveolar epithelial cells (Chan et al., 2013). The MERS-CoV virus is thought to be an animal virus that has sporadically resulted in human infections, with subsequent limited transmission between humans. The evidence for the animal origin of the virus is circumstantial.
Nevertheless, the alternative explanation to explain sporadic appearance of severe human cases with long periods of time between them, and the wide geographical area over which the virus was apparently distributed, is unrecognized ongoing transmission in people. Surveillance efforts, since the discovery of the virus and retrospective testing of stored respiratory specimens suggest this is not the case (Gautret et al., 2013). In August, 2013, the possible source of MERS-COV virus was traced to an Egyptian tomb bat found in a building, in close proximity to an index patient home. The virus genome fragment isolated from the bat was a 100% genetic match to the virus, isolated from the index patient (Jobs, 2013). There had been speculation that, bats might be the source of the virus (Abedine and Saad, 2013). On 9th August, 2013 a report in the journal “The Lancet Infectious Disease” showed that 50 out of 50 (100%) blood serum from Omani camels and 15 out of 105 (14%) from Spanish camels had protein specific antibodies against MERS- COV spike protein and also in Egypt (between 94 and 98% of a total of 110 camels tested positive). A further study on dromedary camels from Saudi Arabia published in December 2013, revealed the presence of MERS–COV in 90% of the evaluated dromedary camels (310), suggesting that dromedary camels not only could be the main reservoir of MERS–COV, but also the animal source of MERS–COV (Hemida et al., 2013).
The Arabian Peninsula remains the site of primary human infection from the zoonotic source of human cases as reported so far and dromedary camels in other parts of North, West and East Africa appear to have serological evidence of MERS infection. The full geographic range of MERS CoV circulation has not been established so far in the above sites including Ethiopia. Sero-prevalence of MERS-CoV in Ethiopia was 97% in adult camels, 93% in young camels and overall prevalence of 96% (Reusken et al., 2013). Therefore the objectives of this study were to determine the seroprevalence of MERS-CoV, to assess the presence of active cases through isolation of MERS-CoV and to identify possible risk factors for the MERS-CoV in the study areas.
Study area and animals
This study was conducted between November 2014 and May 2015 in pastoral areas of Afar (Dubti, Ayssaita, Chifra, Ewa and Amibara districts) and Oromia (Yabello and Fentale districts) regions where camels are populated. Camels of all age and both sexes were randomly selected and included in the study for determination of antibody prevalence against MERS-COV. For the purpose of virus isolation and characterization, nasal swab was taken from 100 camels (Yabelo, Amibara, Fentale and Dubti), 25 from each district.
Geographically, the Afar Regional state is located in the northeastern part of Ethiopia. The total geographical area of the region is about 270,000 km2 (CSA, 2008). It is geographically located between 39°34’ and 42°28’ East Longitude and 8°49’and 14° 30’North Latitude. The region shares common international boundaries with the State of Eritrea in the north-east and Djibouti in the east, as well as regional boundaries with the Regional States of Tigray in the north-west, Amhara in the south-west, Oromia in the south and Somali in the south-east. Afar is characterized by an arid and semi-arid climate with low and erratic rainfall. Rainfall is bi-modal throughout the region with a mean annual rainfall below 500 mm in the semi-arid western escarpments, decreasing to 150 mm in the arid zones to the east. Afar is increasingly in drought prone and the region receives three rainy seasons. The main rain karma accounts for 60% of annual rainfall and occurs from mid-June to mid-September. This is followed by rainy showers in mid-December called dadaa and a minor rainy season during March to April called sugum. Disruptions on the performance of any rainy season will impact on the availability of pasture and water as well as the overall food security situation of the pastoral and agro-pastoral communities (ANRSP, 2010).
Study design and sampling technique
A cross sectional study was conducted for serological investigation of MERS-CoV among dromedary camels found in the study area. Multi stage cluster sampling technique was used to select specific cluster (kebele) for all sites found in the study areas. Finally samples were taken randomly from each selected cluster. Dromedary camels found in Afar and Oromia region of both age groups (young 1 to 3 years and adults > 3 years of age), both sex groups (male and female) and two categories of herd size (large herd size=herd having above 50 camels and small herd size = herd having less than or equal to 50 camels) were included in the study.
Sample size
Based on Thrusfield (2007) formula, the sample size of study animals was calculated.
n = 59 and the sample size multiplied by the cluster number to get the final sample size 59 x 8 sites = 472 (Total sample size required).
But additional 97 samples were taken to increase the precision of the study and the final sample size became 569. Where n = required sample size, Pexp = expected prevalence (96%) and d = desired absolute precision (95%)
Sample collection, transportation and storage
The samples were taken from randomly selected dromedary camels from each cluster (kebeles) of randomly selected Zones and woredas of Afar and Oromia. Then 10 mm of blood sample was collected from jugular veins of the animals using needle and plain test tube (without anticoagulants). The blood was allowed to clot for2 h at room temperature, stored overnight at 4°C and then, serum was separated from the clot standing for 20 min at room temperature, to allow for clot formation.
Serum was separated from the clot by centrifugation at 3000rpm for 10 min and transferred to crayovial tubes. Then the separated serum was labeled and kept under refrigeration (–20°C) until transported to National Veterinary Institute (NVI) for storage, before taken to the University of Hong Kong for analysis.
The collected 569 sera samples were transported in the icebox to National Veterinary Institute serology laboratory and kept under refrigeration (–20°C) until transported to University of Hong Kong for serological analysis. Hundred swabs were collected by using cotton swab from randomly selected sites (Borena and Metehara) from Oromia region (Dubti and Melka werer). Twenty five swabs were taken from each site mentioned above. The swabs were preserved in buffer solution until transported to NVI virology section for deep freezing at -80°C. Finally the swabs transported to Hong Kong University for analysis.
Laboratory analysis
The blood samples collected were analyzed by two serological tests MERS-CoV spike, pseudoparticle neutralization assay (screening test) and Micro neutralization tests (confirmatory test).
The swabs collected undergo extraction of DNA and Polymerase Chain Reaction (PCR) to identify the presence of MERS-CoV in the sample.
Serology
MERS-CoV spike pseudo particle neutralization assay
A codon-optimised spike gene was designed according to published MERS-CoV genome sequence (GenBank accession number: JX869059.1), synthesised by GeneCust (Luxembourg) and subcloned into pcDNA3.1+ vector to generate pcDNA-S. To produce HIV/MERS spike pseudoparticles, 10 μg pNL Luc E- R- and 10 μg pcDNA-SS were co-transfected into 4x106 293T cells. Supernatants of transfected cells were harvested 48 h later and quantified for HIV p24 viral protein using a p24 ELISA Kit (Cell Biolabs, San Diego, United States).
For ppNT assay, HIV/MERS pseudoparticles containing 5 ng p24 were used to infect Vero E6 cells (ATCC CRL-1586) in a single well (96-well plate format; 1x104 cells/well). Infected cells were lysed in 20 μl lysis buffer and 100 μl of luciferase substrate at 2days postinfection (Promega Corporation, Madison, United States).
Luciferase activity was measured in a Microbeta luminometer (PerkinElmer, Waltham, United States). For ppNT, HIV/MERS pseudoparticles (5 ng of p24) were pre-incubated with serially diluted sera for 30 min at 4°C and then added to cells in triplicate. Residual virus replication was assayed at 2days post infection, as described above. The highest serum dilution giving a 90% reduction of luciferase activity was regarded as the ppNT antibody titre.
Viruses and virus titration
MERS-CoV (strain EMC) virus was obtained from Dr R Fouchier (Erasmus University Medical Center, Rotterdam, and the Netherlands). SARS-CoV (strain HKU-39849) was taken from the virus repository at Hong Kong University. Virus stock for MERS-CoV was prepared in Vero cell culture (ATCC CCL-81) in minimal essential medium containing 2% fetal bovine serum, 100 units/mL penicillin and 100 μg/mL Streptomycin. Virus aliquots were stored at -80°C. Virus was titrated in serial half-log10 dilutions (from 0.5 log to 7 log) to obtain 50% tissue culture infectious dose (TCID50) on 96 well tissue culture plates of Vero cells. The plates were observed in a phase contrast microscope for cytopathic effect (CPE) daily, for three days. The endpoint of viral dilution leading to CPE in 50% of inoculated wells was estimated, by using the Reed Muench method and designated as one TCID50. SARS-CoV was grown and titrated in the same manner with the exception that Vero E6 cells (ATCC CRL-1586) were used.
Micro-neutralization tests
Serial two fold dilutions of heat inactivated sera (56°C for 30 min) were made, starting with a dilution of 1:10. The serum dilutionswere mixed with equal volumes of 200 TCID50 of MERS-CoV or SARS-CoV as indicated. After 1 h of incubation at 37°C, 35 μL of the virus serum mixture was added in quadruplicate to Vero or Vero-E6 cell monolayers for MERS-CoV and SARS-CoV, respectively, in 96 well microtiter plates. After 1 h of adsorption, an additional 150 μL of culture medium were added to each well and the plates was incubated for three more days at 37°C in 5% CO2 in a humidified incubator. A virus back-titration was performed without immune serum to assess input virus dose. CPE was read at three days post infection. The highest serum dilution that completely protects the cells from CPE in half of the wells was taken as a neutralizing antibody titre and was estimated using the Reed-Muench method. Positive and negative control sera were included to validate the assay.
Extraction of RNA and Polymerase Chain Reaction (PCR)
Real-time reverse transcription PCR (RT-PCR) targeting upstream of E gene of MERS-CoV was used for screening. The open reading frame (ORF) 1a gene was used for confirmation as recommended by the World Health Organization. We also used a previously described pan-CoV nested PCR, targeting the viral RNA-dependent RNA polymerase (RdRp) region (13), and PCR products were analyzed by sequencing (online Technical Appendix, wwwnc.cdc.gov).EID/article/20/6/14-0299-Techapp1.pdf). We detected MERS-CoV RNA in (n=7/100) (7%) of 100 nasal swab specimens from dromedary camels with the upstream of E gene assay (cycle threshold [Ct] 23.2–36.8), confirmed by the ORF1a assay (Ct 23.2–39.1), fulfilling the World Health Organization criteria for diagnosis of MERS-CoV infection. PCR was repeated from a fresh RNA extract to confirm positive results.
Data analysis
Data obtained from the investigations was coded and stored in Excel spread sheets. The independent variables age, sex, herd size and origin (kebele) of the dromedary camels have been compared to evaluate their effect on the magnitude of the disease. The collected data was analyzed using SPSS version 20 software. Descriptive statistics was used to determine the frequency of proportion (prevalence) and chi square, and logistic regression was used to check the association between the sero-prevalence and the independent variables. The precision level used was 5% with the level of significance differences set at p < 0.05, chi square(x2) > 3.841 for association and logistic regression with odds ratio >1 for the magnitude of the risk(effect) of exposure variables.
Seroprevalence of MERS-CoV
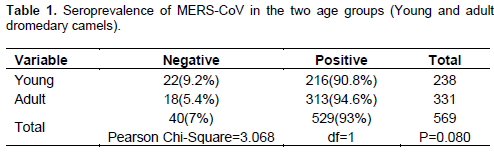
From the total of 569 camel sera tested, 93% (n=529/569) were positive for MERS-COV specific antibody with MERS-CoV spike pseudoparticle neutralization assay and Microneutralisation tests. Out of 238 sera tested from young Dromedary, camels (n=216/238) (90.8%) were positive and from 331 adult, camels (n=313/331) (94.6%) were also positive and the difference seen was not significant (Pearson Chi-Square=3.068 and P=0.080) (Table 1).
From 569 dromedaries camel sampled 361 were Female and 208 were Male. (n=339/361) (93.9%) and (n=190/208) (91.3%) of Female and Male were positive respectively. The difference seen between the two sex groups was insignificant (Pearson Chi-Square=1.323 and P=0. .250) (Table 2).Out of 222 Camels tested from small herd sized group (n=205/222) (92.3%) were positive and from 347 large herd sized camels (n=324/347) (93.4%) were positive but the difference observed was not statistically significant (Pearson Chi-Square=0.220and P=0.639 (Table 3).Out of 230 camels tested from Oromia region (n=216/230) (93.9%) were positive and 339 samples from Afar (n=313/339) (92.3%) were positive but the difference was not statistically significant (Pearson Chi-Square =0.525 and P=0.469) (Table 4).
Among Zones included in the study highest prevalence recorded in East Shoa (n=136/137) (99.3%) and the lowest at Borena (n=80/93) (86.0%).The prevalence difference among Zones was highly statistically significant (Pearson Chi-Square=24.285 and P=0.000) (Table 5). Out of 7 districts included in the study the highest prevalence was observed in Fentale (n=136/137) (99.3%) and the lowest in Yabello (n=80/93) (86.0%) with Pearson Chi-Square=27.173 and P=0.000 value) (Table 6). Among the kebeles (n=136/137) (99.3%) prevalence was the highest prevalence from Adis Ketema of Metehara and the lowest Geriro (n=19/23) (82.6%) of Afar with Pearson Chi-Square=28.308 and P=0.000 values (Table 7).
Molecular identification of MERS-CoV
From the 100 swab randomly sampled (n=7/100) (7%) of them evident circulation of MERS-CoV in the study area 2 from Dubti, 1 from Fentale and 4 from Amibara (Table 8).
The overall high prevalence (n=529/569) (93%) evident in this study completely agree with study done in Ethiopia by (Reusken et al., 2013). The prevalence recorded in Young Dromedary camel (n=216/238) (90.8%) and Adult (n=313/331) (94.6%) was also similar with work of the same author (93% in young Dromedary and 97% in Adult). The higher prevalence observed in the Adult Dromedary camel, may be due to their access to MERS-CoV infected animal at common watering place found in the study area. The overall prevalence of MERS-CoV in this study also agree with study done in Egypt by (Chu et al., 2014), (n=48/52) (92.3%) using the same pseudoparticle neutralization assay as the current study. The slight lower prevalence recorded in Female dromedary camels of this study (n=339/361) (93.9%) compared to the Males (n=190/280) (91.3%) may be due frequent interaction of female camels with humans and their calves.
The two herd size categories in the current study have little variation in their prevalence. Camels in the larger herd size have slightly higher prevalence (n=324/347) (93.4%) than the small herd sized (n=205/222) (92.3%), but the difference between the herds categories was not statistically significant. This may be related with the possibility of increment of an infected animal in the herd which further facilitate transmission of the virus. The prevalence observed in Oromia region was higher (n=216/230) (93.9%) as compared to Afar (n=313/339) (92.3%), this may be because of the high contact rate of the camels in each other during watering in Oromia as compared to the Afar camels. Among the five Zones included in the study, the highest prevalence was evident in East Shoa (n=136/137) (99.3%) and the lowest in Borena zone (n=80/93) (86.0%).The difference observed among the zones was highly significant (p=0.000).
Fentale was the district where the highest prevalence of MERS-CoV recorded (n=136/137) (99.3%) the lowest in Yabelo district (n=80/93) (86.0%) both from Oromia regional state of Ethiopia. Nine kebeles were included in the study with highest prevalence from Adis Ketema (136/137) (99.3%) and the lowest from Geriro (n=19/23) (82.6%).The difference observed was statistically significant (P=0.000). Hundred nasal swabs were used for identification of MERS-CoV from Yabelo, Fentale, Dubti and Amibara district by RT-PCR. Twenty five swabs from each district except Yabelo MERS-CoV was recovered from all the districts sampled. From the overall 7 identified MERS-CoV cases, Amibara district account the highest (n=4/100) (4%) this may relate with the highest seroprevalence recorded in the area (n=94/95) (98.9%).
Chu et al. (2014) reported identification of MERS-CoV (n=4/110 swabs) (3.6%) from dromedary camels of Egypt using RT-PCR, the same molecular technique used in the current study except MERS-CoV strain (variant) was not identified.
There was high seroprevalence of MERS-CoV evident in the study area (Afar=92.3% and Oromia =93.9%), regional states of Ethiopia. The seroprevalence evidence was supported by identification of the virus by RT-PCR, which indicates existence of active infection of MRS-CoV in the study area. Among 569 Dromedary camel sampled, highest prevalence was observed in East Shoa, Fentale district and the lowest in Borena, Yabelo district both from Oromia regional state of Ethiopia.
The study also implies the probability of circulation of MERS-CoV in human being in the study areas, due to the zoonotic nature of the virus and is transmissibility between Dromedary camel and human. Therefore, further study should be conducted to identify which specific factors favor the high prevalence observed, identify the strain (variant) of MERS-CoV in the area and serological and molecular characterization to check the status of the disease in human beings in Ethiopia.
The authors have not declared any conflict of interests.
REFERENCES
|
Abedine, Saad (2013). Death toll from new SARS-like virus climbs to 9. CNN news March 13, 2013 NRSP (2010): Semera, Ethiopia. P 67.
|
|
|
|
Assiri A, McGeer A, Perl TM (2013). Hospital outbreak of Middle East respiratory syndromecoronavirus. N. Engl. J. Med. 369(5):407-416.
Crossref
|
|
|
|
|
Chan RW, Chan MC, Agnihothram S, Chan LL, Kuok DI, Fong JH, GuanY, Poon LL, Baric RS, Nicholls JM, Peiris JS (2013). Tropism of innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J. Virol. 87(12):6604-6614.
Crossref
|
|
|
|
|
Chu DKW, Poon LLM, Gomaa MM, Shehata MM, Perera RA, Zeid DA (2014). MERS coronaviruses in dromedary camels, Egypt. Emerg. Infect. Dis. 20(6):1049-53.
Crossref
|
|
|
|
|
CSA (2008). Agricultural sampling survey report on area and crop production, Addis Ababa.
|
|
|
|
|
Gautret P, Charrel R, Belhouchat K, Drali T, Benkouiten S, Nougairede A, Zandotti C, Memish ZA, al Masri M, Gaillard C, Brouqui P, Parola P, (2013). Lack of nasal carriage of novel coronavirus (HCoV-EMC) in French Hajj pilgrims returning from the Hajj 2012, despite a high rate of respiratory symptoms. 29(7):E315-E317.
|
|
|
|
|
Hemida MG, Perera RA, Wang P (2013). Middle East respiratory syndrome (MERS) coronavirus Seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Eurosurvill.50, 200659.
Crossref
|
|
|
|
|
Marshall K (2014). The Complex and Gender Differentiated Objectives of Livestock Keeping for Somali Pastoralists. Proceedings of the 10th World Congress of Genetics Applied to Livestock Production, Vancouver, Canada.
|
|
|
|
|
Reusken CB (2015). Occupational Exposure to Dromedaries and Risk for MERS-CoV Infection, Qatar, 2013-2014, Emerg. Infect. Dis. 21(8):1422-1425.
Crossref
|
|
|
|
|
Reusken CBEM, Haagmans BL, Muller MA, Ashenafi F, Fufa D, Endrias Z, Lilia M, Hussaini U, Simenew M, Gert-Jan G, Yusuf W, Marion PG (2013). Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 13(10):859-866.
Crossref
|
|
|
|
|
Thrusfield M (2007). Veterinary Epidemiology, 3rd Edition, University of Edinburgh, Scotland UK.
|
|
|
|
|
WHO (2013). Coronavirus infections.
|
|
|
|
|
Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA (2012). Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367(19):1.
Crossref
|
|