ABSTRACT
Mycobacterium avium subspecies paratuberculosis (Map) causes paratuberculosis, an infectious enteritis that affects domestic ruminants. The main source of infection for herds comes from the elimination of the bacilli through feces. The objective of this study was to determine Map excretion in bovine feces. The study included forty, one-year-old bovine, Holstein breed females, from a herd that had >25% paratuberculosis prevalence. Samples of blood and feces were obtained four times with three-month intervals. Feces samples were subjected to bacteriological culture and DNA extraction for IS900 quantitative real time polymerase chain reaction (Q-PCR). Enzyme-linked immunosorbent assay (ELISA) was done from serum samples to detect the presence of anti-Map antibodies. Correlation between IS900 Q-PCR, culture and ELISA was established by the Kappa (K) test; statistical analysis was carried out by the Pearson Chi2 test. On the first sampling, five animals were detected by IS900 Q-PCR, shed 1 × 102 to 2.6 × 103 Map copies per gram of feces; on the second sampling, 6 animals shed 3.25 × 104 to 8.5 × 108; on the third sampling two animals shed 2.03 × 105 to 1.10 × 106 and on the fourth, three animals shed 7.92 × 104 to 1.4 × 107 Map copies. Correlation between tests in samplings first, second and fourth, was 0.53 to 0.73, and for third was 0.22. With the use of IS900 Q-PCR, it was possible to detect animals that were eliminating Map in feces from 12 months of age without clinical manifestations. The IS900 Q-PCR is an alternative method to carry out programs of control of paratuberculosis that allow the detection of animals that shed Map in the early stages of the infection.
Key words: Paratuberculosis, Mycobacterium avium subspecies paratuberculosis, excretion, quantitative polymerase chain reaction (Q-PCR), bacteriological culture, enzyme-linked immunosorbent assay (ELISA).
Paratuberculosisis is a chronic infectious disease, caused by Mycobacterium avium subspecies paratuberculosis (Map) that affects domestic and wild ruminants. Map is classified within the Mycobacterium avium-intracelullare complex (M. avium subspecies avium, M. avium subspecies paratuberculosis, M. avium subspecies silvaticum and M. intracelullare) and it differs from other subspecies of the complex by its dependence on mycobactin for in vitro growth (Stevenson et al., 2002). Map is an intracellular bacteria characterized by resistance to the environmental conditions; it can remain viable for 270 days in stagnant waters, 47 months in dry desiccated organic matter, 45 days in milk and 60 days in fresh cheese (Abalos, 2001; Cirone et al., 2007).
The disease has a chronic course and mainly affects adult animals of two to three years of age. It is characterized by progressive loss of body condition due to the lesions produced in the ileum, ileocecal, cecum and proximal colon mucosa and mesenteric lymph nodes; diffuse hypertrophy of the jejunum and ileum mucosa is developed, which acquire a rough appearance, causing low nutrient absorption (Valentin-Weigand, 2002).
Paratuberculosis has an important impact on animal production by generating a reduction in milk and meat production, early discarding of animals and low fertility with predisposition for mastitis (Eda et al., 2006). With this disease, carcasses have a lower commercial value and there is an increase in control program costs (Soto et al., 2002a). Map is excreted intermittently and animals are infected by consuming feed and water contaminated with paratuberculosis microorganisms shed by infected animals. Map concentration in feces may surpass 108 colony forming units (CFU)/g (Valentin-Weigand, 2002).Young animals less than six months of age are the most susceptible to infection; calves that are in subclinical stages of the disease shed the bacilli in feces without showing the characteristic signs of the disease (Park et al., 2006). The disease distribution is worldwide, especially in domestic ruminants confined rearing conditions; prevalence of the disease varies between 5 and 55% (Milian-Suazo et al., 2015; Avila et al., 2011).
To confirm diagnosis, bacteriological culture is carried out, although its disadvantage is that it is a slow process that needs more than six weeks to have a positive Map culture (Soto et al., 2002b). Currently, there are molecular tests developed to diagnose paratuberculosis and the most used is the polymerase chain reaction (PCR). It had been mainly applied as a qualitative method, but there is also the option to do the PCR test in real time or quantitative PCR (Q-PCR) that is a variation of the standard PCR used for quantification of DNA or messenger RNA (mRNA) of a sample. It is possible to determine the number of copies or the relative amount of a specific DNA or RNA sequence by using specific sequencing primers. The amount of amplicon produced in each PCR cycle is used for quantification. Product quantification occurs by the addition of fluorophores that join the amplicon in a directly proportional manner, and therefore, the higher amount of product there is, the higher fluorescence emission. Real time PCR programs detect the amount of fluorescence produced in each PCR cycle and the analysis programs represented the said fluorescence in graphs, in relation to the number of cycles (UCV, 2012; Vinueza-Burgos, 2009). Most of the PCR protocols, for DNA/RNA Map detection, use primers that amplify the 900-insertion sequence (IS900), and therefore it is been accepted as a standard Map marker. IS900 is a 1451 base-pair sequence that codifies for a hypothetical transposase of 399 amino acids, with 14 to 20 copies in the MAP genome inserted in a consensus region within highly conserved loci in the Map genome (Kralik et al., 2011; Bull et al., 2000). It is thought that, for every animal diagnosed with the clinical profile of paratuberculosis, there are approximately 25 other individuals that have a subclinical profile and that these will intermittently shed the bacillus through feces. It is therefore important that the diagnosis of this disease be done using diagnostic techniques, such as q-PCR, that can detect animals that are shedding the bacillus before clinical signs are present, in order to establish control measures within the herd. The purpose of this study was to determine the shedding of Map in bovine feces using Q-PCR.
The work was carried out using 40 Holstein breed female cows at least 1-year-old that originated from a dairy herd with previous clinical history of paratuberculosis (>25% prevalence) in Hidalgo Mexico, confirmed with microbiological culture IS900 nested PCR and ELISA. Follow up was done by taking four samples of blood and feces, every three months. Blood was obtained from the caudal vein using vacutainer tubes and centrifuged at 180 × g for 10 min; the resulting serum was then divided into aliquots in microtubes. Feces were collected directly from the rectum using palpation gloves. All samples were stored at -20ºC until analyzed (Martínez et al., 2012).
DNA extraction
Two grams of feces were decontaminated using 50 ml of 7.6% 1-hexadecylpyridinium chloride (HCP) (Sigma Aldrich) for 18 h. DNA extraction was then carried out following the protocol described by Jaimes et al. (2008). DNA was stored at -20°C until used in q-PCR.
Real time Q-PCR
The Q-PCR reaction was done using the primers for ISIS IS900 PTB FP 5´AATGACGGTTACGGAGGTGGT3´ and PTB RP 5´GCAGTAATGGTCGGCCTTACC 3´ (Kim et al., 2002). Each 25-μl q-PCR reaction contained 3 μl DNA (45 ng/μl), 12.5 μl Maxima SYBR green / ROX q-PCR Master mix (3 mM MgCl2, 200 μM each dATP, dGTP, dCTP, 400 μM dUTP, 1.25 IU DNA polymerase and 0.5 IU Uracil-N-Glycosylase (UNG) (Thermo)), 1 μl each PTBFP and PTBRP primers (0.05 μM each/one), and 7.5 μl water. DNA from a standard Map strain (ATCC #43545) was used as positive control, while distilled water was used as the negative control.
The Q-PCR was carried out in a SmartCycler ® (Cepheid) with the following cycling conditions: 95°C/10 min, 40 cycles of 95°C/15 s, 60°C/30 s and 72°C/10 s, followed by a final cycle of 72°C/30 s. Samples were worked in duplicate and the results were analyzed following the guidelines in the SmartCycler operating manual. A linear regression was performed on CT versus log IS900 copy number and R2 was 0.99. The standard error of y was used to create two equations to estimate the upper and lower concentration, for calculation of M. avium subsp. paratuberculosis IS900 copy number; this was done following the protocol described previously by Moravkova et al. (2012), Slana et al. (2008) and Kim et al. (2002).
Bacteriological culture
Feces from all animals were cultured in duplicate following the technique described by Aly et al. (2010). All samples were decontaminated using 0.5% Zephiran and sowns in Herrold’s egg yolk solid media with and without 2 mg/L mycobactin J (Allied Monitor INC) and incubated at 37°C for 6 months. Ziehl-Neelsen acid-fast stain was carried out on cultures that showed positive growth; DNA was also extracted and nested PCR carried out using primers that amplify a 210 bp region of IS900 (Jaimes et al., 2008).
Enzyme linked immunosorbent assay (ELISA)
Protoplasmic antigen of the 3065 Map strain was adhered to ELISA microplates following the protocol described by Martínez et al. (2012). Sera were diluted to a working concentration of 1:160 using a solution of 0.02% Mycobacterium phlei (Allied Monitor INC). A Map positive serum was used as positive control and a negative serum as negative control (Allied Monitor INC). The bovine anti-IgG conjugate tagged with horseradish peroxidase (Sigma-Aldrich) was used at a 1:2000 dilution, while 2, 2'-Azino-Bis-3-Ethylbenzothiazoline-6-Sulfonic acid (ABTS, AMPRESCO) was used as substrate, and 1.5 mM sodium nitrate (sigma-Aldrich) in 0.1 M citric acid was used as stop solution. Plate reading was done at 650 nm using a spectrophotometer (ELx800, BioTek). The cut-off was established at 0.22 optical deviations (OD).
Statistical analysis
Statistical analysis was carried out using the STATA® 7.0 software package (StataCorp LP, College Station, TX, USA). Cohen’s Kappa test (k) or inter-rater agreement index was used to establish the association between the results obtained with the IS900 Q-PCR, culture and ELISA. Differences between diagnostic techniques were tested using Pearson’s Chi2 (c2). The scale proposed by Prieto et al. (2014), was used to determine the concordance among techniques. Range (%) to <0.000 is poor; to 0.00 and 0.20 is slight; to 0.21 and 0.40 is fair; to 0.41 and 0.60 is moderate; to 0.61 and 0.80 is substantial; to 0.81 and 1.0 is perfect.
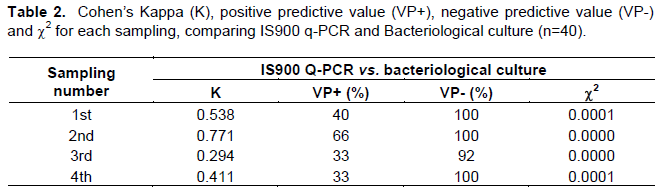
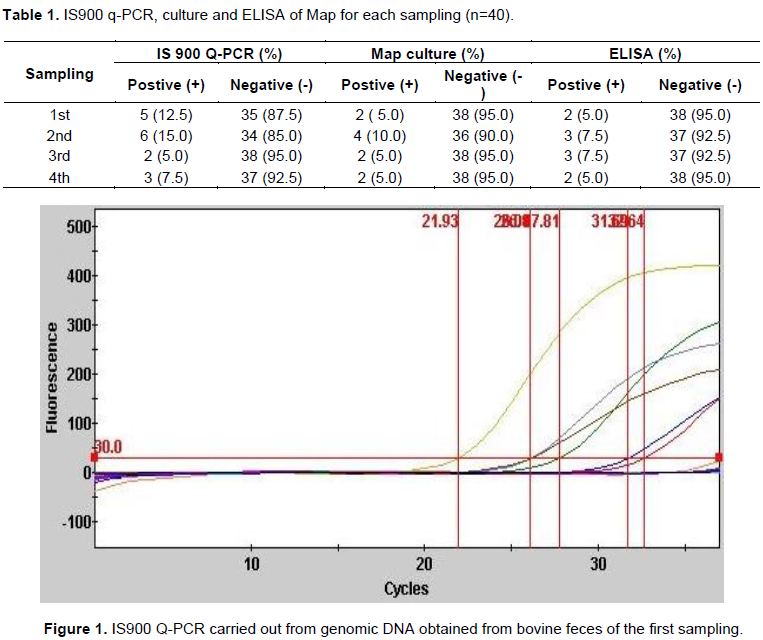
Follow-up of the 40 female cows was done every three months starting at 12 months of age and continued until they reached 21 months, to determine the age at which the animals begin shedding the bacilli in feces. All cows came from a herd with a history of >25% prevalence of paratuberculosis. The results obtained in the work are shown in Table 1, with the use of IS900 Q-PCR, amplification curves of the Map positive control and samples that were found to be positive were present from cycle 20 up to cycle 30 of the Q-PCR run (Figure 1). Animals that were shedding the bacteria could be identified from the first sampling, which corresponded to 12 months of age. Map shedding in feces is intermittent, since only one animal was detected as positive in all samplings.
The presence of anti-Map antibodies was lower (2.5 to 7.5%); tests based on the detection of humoral immune response are not feasible during early stages of infection (subclinical stage), because the concentration of anti-Map antibodies in early stages of infection are low.
Bacterial growth could only be observed in tubes that had mycobactin added from five weeks on. Ziehl-Neelsen stain revealed the presence of acid-fast bacilli, and the nested PCR amplified the expected 210 bp amplicon which is specific to Map. The sensitvity of the bacteriological culture was less than 10%, the animals that were positive to the bacteriological culture in all the samplings were also positive to the IS900 Q-PCR.
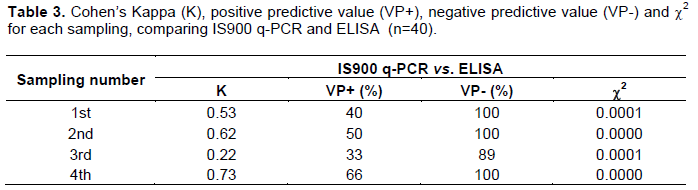
Association between the three diagnostic tests used in this study was determined with Cohen’s Kappa test (Tables 2 and 3). When comparing IS900 Q-PCR with bacteriological culture and ELISA, first, second and fourth samplings showed moderate to substantial concordance, while in third sampling the concordance was fair.
Results of the negative predictive values (VP-) indicate that IS900 Q-PCR is a highly specific test since the average VP-reached 98% when compared with the microbiological culture and 97.5% when compared with ELISA. As such, individuals that had a negative result in the IS900 Q-PCR test had been correctly diagnosed as they were true negatives. The positive predictive value (VP+) reached 43 and 47.5% when comparing IS900 Q-PCR with the microbiological culture and ELISA, respectively (Tables 2 and 3).
Quantification of Map shedding
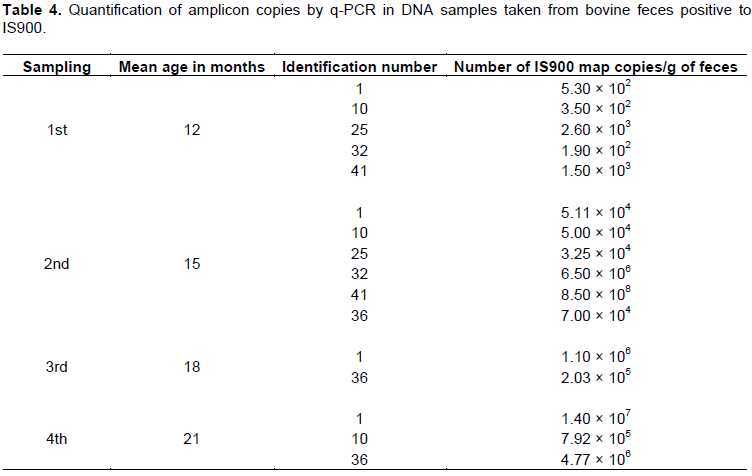
Map shedding is intermittent and starts in the early stages of the infection. The IS900 Q-PCR detected bovine from 12 months of age, that were already excreting the bacillus in feces, and as the samplings advanced, the elimination increased logarithmically (bovine 1, 10 and 36). On the first sampling, it was detected that five animals shed between 1 × 102 and 2.6 × 103 copies of the IS900 from Map per gram of feces. In the second sampling, six animals shed between 3.25 × 104 and 8.5 × 108. On the third sampling, two animals were detected that shed between 2.03 × 105 and 1.10 × 106, while in the fourth sampling three animals shed between 7.92 × 104 and 1.4 × 107 (Table 4).
Forty dairy cows, which had a mean of 12 months of age at the first sampling, were followed for a year in order to determine the time of shedding of Map using IS900 Q-PCR and bacteriological culture. From the first sampling, animals that shed Map were detected, demonstrating that bovines infected with paratuberculosis can shed the bacillus even in early infection stages without any clinical signs of the disease (Park et al., 2016; Fang et al., 2002).
Previously, Khol et al. (2010), had followed for 380 days a 2.5 year-old cow that had become naturally infected with Map, sampling feces on 9 occasions. Their results showed that the number of IS900 copies that were detected increased between 105 and 107 copies per gram of feces throughout the study, although in their third sample a slight decrease was observed, demonstrating that Map shedding is intermittent and can start from early infection stages (subclinical phase) before the infected animal becomes positive to the serological test. The results agree with those of Khol et al. (2010) in that Map shedding was detected from 12 months of age and that as time advances shedding increased logarithmically from 102 to 106 copies of IS900 of Map (bovines 1, 10 and 25; Table 4). Kralik et al. (2011) developed a predictive model to determine the shedding of Map using Q-PCR with DNA extracted from feces, establishing that the minimum detectable shedding is 102 copies of IS900 of Map per gram of feces. The results obtained agree with those of kralik et al. (2011), because in the first sampling three animals were detected as positive and the quantification was obtained as 1.9 to 5.30 × 102 copies of IS900 of Map per gram of feces.
Several authors have carried out studies on the correlation of q-PCR tests, bacteriological culture and ELISA with the diagnosis of paratuberculosis, with varying results. Pinedo et al. (2008) found that when comparing PCR tests with bacteriological culture and ELISA, Kappa values were 0.39 and 0.01, respectively. In contrast, Fang et al. (2002), when correlating IS900 Q-PCR with bacteriological culture, a Kappa value of 0.94 was found. In this study, Kappa values in the first, second and fourth samplings ranged from 0.53 to 0.73 indicating a moderate correlation, although the Kappa value in third sampling reached 0.22 indicating a fair correlation. It is possible that the latter is due to the fact that in this disease shedding of bacilli is intermittent and at the time of collecting the sample, it could have been absent from feces. Nevertheless, mistakes in DNA extraction or sample contamination could also explain the results and it cannot be excluded (Khol et al., 2010). Correlation between bacteriological culture and Q-PCR depends on the stage of the infection present in the individual at the time of sample collection which has an effect on the amount of bacilli shed (Kralik et al., 2011; Aly et al., 2010; Pinedo et al., 2008). The ELISA used in the study detected less number of animals as positive, this is because, in early infection stages the predominant immune response is cell type, and the elimination of bacillus in feces begins in an intermediate way. Other reason for which the antibody production may be affected, is the cause of loss of immune response after infection. This suppressor activity is known as anergia. Animals in early infection stages, even if they excrete the bacteria in their feces, may not necessarily be detected by the ELISA test, since it may take several months, or years, before the level of circulating antibodies is sufficient to trigger a positive reaction (Soto et al., 2002a).
Bacteriological culture from tissues, feces, milk, and semen, among others, is considered the gold standard for the diagnosis of paratuberculosis. Nevertheless, such testing has its limitations since, although it has a 100% specificity, it has very low sensitivity mainly due to the intermittent nature of the shedding of bacilli through feces, milk and semen (Khol et al., 2010). Furthermore, the culture media needs to be enriched by mycobactin to improve the development of the bacillus and the incubation times are more than six-weeks long for isolating Map. Another important disadvantage of bacteriological culture is the high risk of contamination with other microorganisms, especially fungi, and the decrease in viable bacilli after the sample decontamination process. By using Q-PCR, results are obtained within at least two days, depending on the amount of samples to be processed, representing a clear advantage in time-to-diagnosis when compared with bacteriological isolation allowing the quantification of bacilli shedding in feces (Kralik et al., 2011; Khol et al., 2010). With the simultaneous use of IS900 Q-PCR, fecal culture, and the ELISA test, it is possible to detect a greater number of infected animals.
In control and diagnostic testing, the probability that an individual that has a positive result is indeed positive is known as the positive predictive value, while the negative predictive value is the percentage of individuals with a negative test that do not have the disease. It is important to note that the predictive value is closely related with the sensitivity and specificity of a test, as well as with the prevalence of the disease in the population where the test is to be carried out (Martínez et al., 2012). The positive predictive values (VP+) obtained in this study, ranging between 43 and 47.5%, are considered low and are probably due to the long sampling interval (90 days) and the intermittent shedding of bacilli. Nevertheless, the negative predictive values (VP-) where high (97.5-98%) which demonstrates that the IS900 q-PCR is highly specific and that negative tests were appropriately classified.
The IS900 marker is considered the standard for endpoint PCR and Q-PCR since there are between 14 and 20 copies of the sequence within the mycobacterium genome improving the sensitivity for detecting Map. Other genetic markers have been evaluated for detecting Map, such as the SF57, ISMav2 and HspX sequences, but they have the limitation that they are only present in 1 to 6 copies within the Map genome, therefore sensitivity is reduced, although specificity is increased. To increase specificity when using primers for IS900, these should be designed so that they amplify regions that are close to the 5’ end of this inserted sequence since it is highly conserved (Cook and Britt, 2007; Tasara and Stephan, 2005; Kim et al., 2002; Fang et al., 2002).
In this study, the IS900 Q-PCR detected that 12.5% of animals of the 12 months of age that were included in the first sampling where shedding bacilli through feces. This is important since the majority of the paratuberculosis clinical cases are observed in animals with two to six years of age and young animals, in general, do not show clinical signs but are already shedding the bacillus through their feces. As such, it is important that diagnostic tools are available that can detect the shedding of bacilli in the subclinical stage of the disease. In this sense, the IS900 Q-PCR is a good diagnostic alternative for detecting animals that are shedding the pathogen and prevent the spread of the disease by allowing the implementation of control and eradication measures for paratuberculosis.
The IS900 Q-PCR allows the diagnosis of early-stage paratuberculosis in animals from 12 months of age that do not have clinical signology of paratuberculosis. The technique is an alternative to carry out programs of control of paratuberculosis that allow the detection of animals that eliminate Map in early stages of infection.
The authors have not declared any conflict of interests.
REFERENCES
|
Abalos P (2001). News in Paratuberculosis Tecnovet. 7(3). Available at:
View
|
|
|
|
Aly SS, Mangold BL, Whitlock RH, Sweeney RW, Anderson RJ, Jiang J, Schukken YH, Hovingh E, Wolfgang D, Kessel JAS, Karns JS, Lombard E, Smith JM, Gardner IA (2010). Correlation between Herrold egg yolk medium culture and real-time quantitative polymerase chain reaction results for Mycobacterium avium subspecies paratuberculosis in pooled fecal and environmental samples. J. Vet. Diagnos. Invest. 22:677-683.
Crossref
|
|
|
|
|
Avila GJ, Blando GE, Cruz HGE (2011). Paratuberculosis.
|
|
|
|
|
Bull TJ, Harmon-Taylor J, Pavlik I, Zaatari E, Tizard M (2000). Characterization of IS900 loci in Mycobacterium avium subspecies paratuberculosis and development of multiplex PCR typing. Microbiology 146:2185-2197.
Crossref
|
|
|
|
|
Cirone K, Morsella C, Romano M, Paolicchi F (2007). Mycobacterium avium subsp. paratuberculosis in food and its relationship with Crohn's disease. Rev. Argent. Microbiol. 39:57-68.
|
|
|
|
|
Cook KL, Britt JS (2007). Optimization of methods for detecting Mycobacterium avium subspecies paratuberculosis in environmental samples using quantitative, real-time PCR. J. Microbiol. Method 69:154-160.
Crossref
|
|
|
|
|
Eda S, Bannantine JP, Waters WR, Mori Y, Whitlock RH, Scott MC, Speer CA (2006). A Highly sensitive and subspecies-specific surface antigen enzyme-linked immunosorbent assay for diagnosis of Johne’s Disease. Clin. Vaccine Immunol. 13(8):837-844.
Crossref
|
|
|
|
|
Fang Y, Wu W-H, Pepper JL, Larsen JL, Marras SAE, Nelson EA, Epperson WB, Christopher-Hennings J (2002). Comparison of real-time, quantitative PCR with molecular beacons to nested PCR and culture methods for Detection of Mycobacterium avium subspecies paratuberculosisin bovine fecal samples. J. Clin. Microbiol. 40(1):287-291.
Crossref
|
|
|
|
|
Jaimes NG, Santillán FMA, Hernández COA, Córdova LD, Guzmán RCC, Arellano RB, Díaz AE, Tenorio GVR, Cuellar OA (2008). Detection of Mycobacterium avium subspecies paratuberculosis by nested-PCR of ovine fecal samples. Rev. Vet. Méx. 39(4):377-386.
|
|
|
|
|
Khol JL, Kralik P, Slana I, Beran V, Aurich C, Baumgartner W, Pavlik I (2010). Consecutive excretion of Mycobacterium avium subspecies paratuberculosis in semen of a breeding bull compared to the distribution in feces, tissue and blood by IS900 and F57 quantitative real-time PCR and culture examinations. J. Vet. Med. Sci. 72(10):1283-1288.
Crossref
|
|
|
|
|
Kim SG, Shin SJ, Jacobson RH, Miller LJ, Harpending PR, Stehman SM, Rossiter CA, Lein DA (2002). Development and application of quantitative polymerase chain reaction assay based on the ABI 7700 system (Taqman) for detection and quantification of Mycobacterium avium subspecies paratuberculosis. J. Vet. Diagnos. Invest. 14:126-131.
Crossref
|
|
|
|
|
Kralik P, Slana I, Kralova A, Babak V, Whitlock RH, Pavlik I (2011). Devolopment of predictive model for detection of Mycobacteiumavium subspeciesparatuberculosis in faeces by quantitative real time PCR. Vet. Microbiol. 149:133-138
Crossref
|
|
|
|
|
Martínez CAG, Santillán FMA, Guzmán RCC, Favila HLC, Córdova LD, Díaz AE, Hernández AL, Blanco OM (2012). Development of an enzyme-linked immunosorbent assay (ELISA) for the diagnosis of bovine paratuberculosis. Rev. Mex. Cienc. Pecu. 3(1):1-18.
|
|
|
|
|
Milian-Suazo F, Santillán-Flores MA, Zendejas-Martínez H, García-Casanova L, Hernández-Andrade L, Canto-Alarcón GA (2015). Prevalence and associated risk factors for Mycobacterium avium subspecies paratuberculosis in cattle in México. J. Vet. Med. Anim. Health 7(10):302-307.
Crossref
|
|
|
|
|
Moravkova MV, Babak V, Kralova A, Pavlik I, Slana I (2012). Culture- and quantitative IS900 real-time PCR-based analysis of the persistence of Mycobacterium aviumsubspecieparatuberculosis in a controlled dairy cow farm environment. Appl. Environ. Microbiol. 78(18):6608-6614.
Crossref
|
|
|
|
|
Park HT, Shin MS, Park HE, Cho YI, Yoo HA (2016). PCR-based detection of Mycobacterium avium subspecies paratuberculosis infection in cattle in South Korea using fecal samples. J. Vet. Med. Sci. 78(9):1537-1540.
Crossref
|
|
|
|
|
Park KT, Ahn J, Davis WC, Koo HC, Kwon NH, Jung WK, Kim JM, Hong SK, Park YH (2006). Analysis of the seroprevalence of bovine paratuberculosis and the application of modified absorbed ELISA to field sample testing in Korea. J. Vet. Sci.7(4):349-354.
Crossref
|
|
|
|
|
Pinedo PJ, Rae DO, Willians JE, Donovan JE, Melendez P, Buergelt CO (2008). Association among results of serum ELISA, fecal culture and nested PCR on milk, blood and feces for detection of paratuberculosis in dairy cows. Transbound. Emerg. Dis. 55:125-133.
Crossref
|
|
|
|
|
Prieto A, Lago N, Díaz JM, Pérez I, Guarddon M, Díaz P, López C, Fernández G (2014). Diagnosis of bovine paratuberculosis: Comparison between fecal culture and Q-PCR on field samples. XIX International Congress ANEMBE of bovine medicine. Ovideo, Spain, pp. 65-70
|
|
|
|
|
Slana I, Kralik P, Kralova A, Pavlik I (2008). On-farm spread of Mycobacterium aviumsubspeciesparatuberculosis in raw milk studied by IS900 and F57 competitive real time quantitative PCR and culture examination. Int. J. Food Microbiol. 128:250-257.
Crossref
|
|
|
|
|
Soto JP, Kruze J, Leiva S (2002a). Isolation of Mycobacteriumaviums subspecies paratuberculosis of faeces in dairy herds infected by the modified Cornell method. Arch. Med. Vet. 34(2):1-5
|
|
|
|
|
Soto JP, Kruze J, Leiva S (2002b). Comparison of three different methods for the diagnosis of bovine paratuberculosis in infected dairy herds. Arch. Med. Vet. 34(2):10-15
|
|
|
|
|
Stevenson K, Hughes VM, de Juan L, Inglis NF, Wright F (2002). Molecular characterization of pigmented and nonpigmented isolates of Mycobacterium aviumsubspecie paratuberculosis. J. Clin. Microbiol. 40:1798-1804.
Crossref
|
|
|
|
|
Tasara T, Stephan R (2005). Development of an F57 sequence-based real-time PCR assay for detection of Mycobacterium avium subspecie paratuberculosis in milk. Appl. Environ. Microbiol. 71(10):5957-5968.
Crossref
|
|
|
|
|
University of the Vasco Country (UVC) (2012). Análisis de expresión génica mediante PCR a tiempo real o Q-PCR.
|
|
|
|
|
Valentin-Weigand P (2002). Johne's Disease: Pathogenesis and Problems Related to Diagnosis. In Recent Developments and Perspectives in Bovine Medicine. XXII World Buiatrics Congress, Hanover, pp. 18-23.
|
|
|
|
|
Vinueza-Burgos C (2009). Real Time PCR: The new era of celular genetic information. Rev. Elect. Vet. 10(2):1-13.
|
|