ABSTRACT
Saffron (Crocus sativus L.) is a triploid sterile plant characterized by its long red stigmas, which produce and store its chief carotenoid derivatives-safranal, crocin and picrocrocin. Saffron selections of Kashmir are heterogeneous for floral characteristics which are mainly attributed to the environmental factors, though genetic factors may have role with regard to its differential characteristics across various selections found in the area. Identification of high yielding selections using the existing gene pool of saffron may help in improving the productivity of this crop. The present study was conducted at Biotechnology Lab, Central Institute of Temperate Horticulture, Srinagar, India during 2012-2013 to find out the apocarotenoid gene expression in thirty-one morphogically distinct clones of saffron. Comparative apocarotenoid gene expression through Real Time PCR analysis revealed a significant variation in Zeaxanthin cleavage dioxygenase (CsZCD) and lycopene-β-cyclase (CsLYC) genes between the most divergent selections (CITH-S-107 and PAM-S-116) indicating a possible role of these genes for regulating the apocarotenoid production in stigma. Significant variation was observed with respect to stigma length (2.86 to 4.84 cm) across thirty-one selected saffron clones. Although some clones showed variation with respect to stigma number, in addition to normal trifid stigmas, some clones produce tetrafid stigmas as well but this character was not heritable. Our study provides sufficient knowledge to identify clones with better stigma characteristics and higher apocarotenoid biosynthetic potential for further crop improvement programs.
Key words: Crocus sativus, CsZCD, CsLYC, RT-PCR, RNA isolation, apocarotenoid.
Crocus sativus unknown as a wild plant is considered to be a mutant that has derived from C. cartwrightianus. The cultivated clone was probably selected for its triploid vigour and extra long stigmas and has been maintained in cultivation for over 3000 years. The dried red stigmas of C. sativus, has been used as flavouring and colouring agent since then and is currently considered the world’s most expensive spice. The major components of saffron are the apocarotenoids cis- and trans-crocins, picrocrocin (β-D-glucopyranoside of hydroxyl-β-cyclocitral), and its degradation product, the odour-active safranal (Kanakis et al., 2004; Sanchez and Winterhalter, 2013). Clonal selection plays an important role with reference to improving the traits of cultivated saffron. Both experimental mutagenesis and procedures aimed at doubling a chromosome set of saffron have not lead to encouraging results (Nehvi et al., 2007a, b). Therefore, superior clones showing better stigma characteristics viz., higher stigma length, increased number of stigmas and higher apocarotenoid biosynthetic potential needs to be selected and mass multiplied. Furthermore, such genotypes need to be analyzed in detail to find out the active principal behind their superiority, which can be exploited for saffron crop improvement. Apocarotenoids have been extensively studied in the stigma tissue because of their organoleptic properties. It has been proposed that the biogenesis of the main colour principles, crocins, and the odour active compound, safranal is derived by bio-oxidative cleavage of zeaxanthin (Pfander and Schurtenberger, 1982) by a 7, 8 (7’, 8’) cleavage reaction. Apocarotenoid biosynthetic pathway possesses many enzymes which are catalyzing the reactions and coded by related key genes such as PSY, LYC, CCD, BCH and ZCD. Βeta-carotene with two rings is built up via cyclization of lycopene with lycopene-β-cyclase (LYC). The hydroxylation of β-carotene in MVA pathway is catalyzed by β-carotenoid hydroxylase that coded by BCH gene to yield zeaxanthin (Castillo et al., 2005). The biogenesis of the color and odor active compounds of saffron are derived by bio-oxidative cleavage of zeaxanthin (Rubio-Moraga et al., 2009; Gomez-Gomez et al., 2010) at the points 7, 8 (7', 8') by zeaxanthin cleavage dioxygenase (CsZCD) to produce crocetin dialdehyde and picrocrocin. In C. sativus stigmas, the final step involves glucosylation of the generated zeaxanthin cleavage products by glucosyltransferase 2 enzyme which is coded by CsUGT2 gene in chromoplast of stigmas and then sequestered into the central vacuole of the fully developed stigmas (Bouvier et al., 2003).
Thirty-one (31) different saffron clones representing the core collection for saffron germplasm conservation were used in the study. These clones were maintained at Central Institute of Temperate Horticulture, Srinagar (J&K), India. Stigmas were collected from September to November 2013. Freshly cut stigmas were quickly immersed in liquid nitrogen and then stored at -80°C for RNA isolation, while dried stigmas were used for apocarotenoid extraction.
Morphological parameter
The stigma length was measured by digital caliper. Data on stigma length are means of five replicates of 10 flowers. The data was analyzed by comparing means using one way ANOVA and the significance was determined by Duncan’s Multiple Range Test using SPSS for windows (v. 15. SPSS Inc USA).
RNA extraction and cDNA preparation
Frozen stigmas were ground in cold and sterilized mortar and pestle into fine powder and total RNA was extracted using RNA isolation kit (Roche Applied Science “Penzberg, Germany”) following the manufacturer's protocol. Quality of the extracted RNAs was checked by measuring the absorbance at 260 and 280 nm using Nano-Drop and RNAs with ratio of OD 260/280 ranging from 1.2 to 1.5 were used for cDNA synthesis. For first-strand cDNA synthesis, 5 µg of total RNA template and 18-bp oligo dT primer and cDNA synthesis kit (Roche Applied Science “Penzberg, Germany”) were used as described by the manufacturer. The cDNA was stored at -20°C until use.
The concentration of RNA was determined using the Beer-Lambert law, which predicts a linear change in absorbance with concentration. An A260 reading of 1.0 is equivalent to about 40 µg/ml of RNA and the OD at 260 nm is used to determine the RNA concentration in a solution. RNA has its absorption maximum at 260 nm and the ratio of the absorbance at 260 and 280 nm is used to assess the RNA purity of an RNA preparation. Pure RNA has an A260/A280 of 2.1. The NanoDrop® ND-1000 UV-Vis Spectropho-tometer enables highly accurate analyses of extremely small samples with remarkable reproducibility. The sample retention system eliminates the need for cuvettes and capillaries, which decrease the amount of sample, required for the measurement.For apocarotenoid extraction, saffron stigmas were extracted with methanol (100 mL) in a microcentrifuge tube for 5 min on ice. Tris-HCl (50 mM, pH 7.5; containing 1 M NaCl) was then added (100 mL) and incubated for 10 min on ice. The precipitate was collected by centrifugation at 3,000 g for 5 min at 4°C. The pellet was then reground in acetone (400 mL) and incubated on ice for 10 min. The mixture was centrifuged at 3,000 g for 5 min at 4°C. This step was repeated until no color was detected in the pellet. The supernatants were pooled and evaporated and the dried residues were stored at -80°C.
RT- PCR analysis
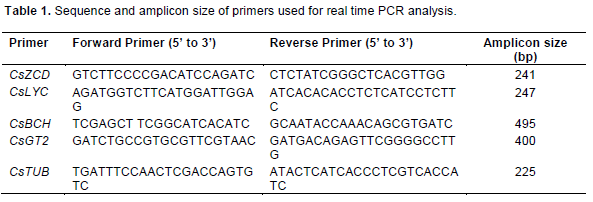
Reverse transcription was carried for amplification of CsZCD and CsLYC with CsTUB gene as internal control, using AMVRT cDNA kit (Roche Applied Science, “Penzberg, Germany”) according to user manual. RNA isolated from all the samples was quantified spectrophotometrically and equal concentration of RNA was used for semi-quantitative comparison of gene expression through reverse transcription polymerase chain reaction. RNA was handled under RNase free environment and all operations were carried out using -20°C cooler. Forward and reverse primers sequences used for amplification of these genes and expected length of amplicons are shown in Table 1. PCR reactions were performed in thermocycler (Takara, Japan) with 2 to 5 µg of cDNA. Initial denaturizing at 95°C for 5 min followed by 35 cycles of amplification according to the subsequent scheme; denaturizing 1 min at 94°C, annealing at 56.2°C for 30 s and extension at 72°C for 40 s and final extension at 72°C for 7 min. The experiments were repeated twice. Subsequently 5 µl of the PCR products were used on 1.2% (w/v) agarose (Sigma-Aldrich, St Louis, MO, USA).
Real time PCR analysis
The Real Time PCR was performed in 96-well plates with a LightCycler 480 real-time PCR instrument (Roche Diagnostics) using the LightCycler 480 SYBR Green I Master kit. SYBR Green I is a DNA double-strand-specific dye. During each phase of DNA synthesis, the SYBR Green I dye, which is included in the reaction mix, binds to the amplified PCR products; the amplicon can be detected by its fluorescence. Hot start protocols used with the LightCycler 480 SYBR Green I Master have been shown to significantly improve the specificity, sensitivity, and yield of PCR. Heat-labile blocking groups on some of the amino acid residues of FastStart Taq DNA Polymerase make the modified enzyme inactive at room temperature. Therefore, there is no elongation during the period when primers can nonspecifically bind. The FastStart Taq DNA Polymerase is “activated” by removing the blocking groups at a high temperature (that is, a pre-incubation step at +95°C for 5 min).
The LightCycler® 480 SYBR Green I Master is a one-component hot start reaction mix for PCR. It contains FastStart Taq DNA Polymerase and DNA double-strand-specific SYBR Green I dye for product detection and characterization. Since the mix is provided as an easy-to-use real master reagent, reaction setup only requires the addition of template DNA and primers. The mix can be used with different types of DNA (e.g., genomic, cDNA) and is ideally suited for high-throughput applications in 96- or 384-well plates. The LightCycler® 480 SYBR Green I Master is a one-component hot start reaction mix for PCR. It contains FastStart Taq DNA Polymerase and DNA double-strand-specific SYBR Green I dye for product detection and characterization. Since the mix is provided as an easy-to-use real master reagent, reaction setup only requires the addition of template DNA and primers. The mix can be used with different types of DNA (e.g., genomic, cDNA) and is ideally suited for high-throughput applications in 96- or 384-well plates.Reactions were performed in triplicate and contained 5 μl SYBR Green I Master, 2 μl PCR-grade water, 2 μl cDNA, and 0.5 μl of each of the 10 μM forward and reverse gene-specific primers in a final volume of 10 μl. The reactions were incubated at 95°C for 5 min, followed by 40 cycles of 95°C for 15 s, 56.2°C for 15 s, and 72°C for 20 s. In our study reporter system, the intercalating SYBR green assay was used. SYBR green binds to all double-stranded DNA via intercalation between adjacent base pairs. When bound to DNA, a fluorescent signal is emitted following light excitation.
As amplicon numbers accumulate after each PCR cycle, there is a corresponding increase in fluorescence. Post-PCR dissociation (melting) curve analysis (60 to 95°C) was carried out to confirm that the fluorescence signal is generated only from target templates and not from the formation of nonspecific PCR products. LightCycler 480 software (version 1.5; Roche Diagnostics) was used to collect the fluorescence data. Advanced relative quantification between the genotypes and three stages of stigma development were done through 2-??CT method (Livak and Schmittgen, 2001).
Average stigma length varies from 2.86 cm in selection PAM-S-116 to 4.84 cm in CITH-S-107 (Table 2), which is higher than reported earlier by Caiola (2004) and Nehvi et al. (2007a). Nehvi et al. (2007a) reported stigma length range of 2.41 to 3.87 in 438 random selections of saffron from Kashmir. Stigma length variation was successfully transferred to next generation; hence this is a stable inherited trait hence can be tagged with molecular markers for association studies. The results are in accordance with the erlier reports of Mir et al. (2012b). Genetic variation and heritability of agro-morphological and phytochemical traits in saffron populations have been studied, populations were found significantly different for most evaluated traits like leaf number per plant, leaf length, fresh stigma characteristics, dry stigma weight per plot, spathe number and the content of crocins, picrocrocin and safranal (Baghalian et al., 2010).
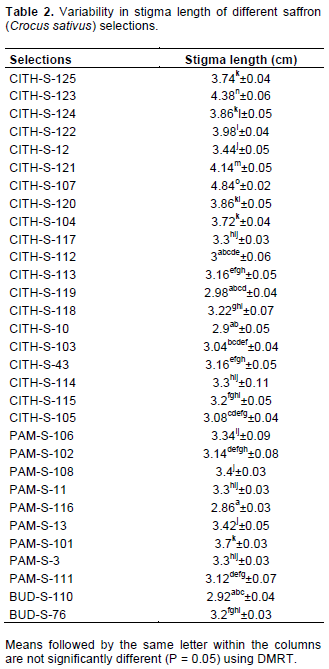
RT-PCR for semi quantitative expression of CsZCD and CsLYC genes between two distinct genotypes (CITH-S-107 and PAM-S-116) was done during scarlet stage of stigma development. CsTUB was used as an internal control. These genotypes did not show significant variation, however, CsZCD gene showed higher expression than CsLYC gene (Figure 1). Although, reverse transcription PCR analysis is the semi-quantitative method of gene expression but it is very good method to find an idea about comparative level of gene expression and is a highly sensitive and specific method useful for the detection of rare transcripts or for the analysis of samples available in limiting amounts. RT-PCR is increasingly used to detect small changes in gene expression that would otherwise be undetectable (Freeman et al., 1999).
Quantitative real time PCR (Q-PCR) approaches combine the detection of target template with quantification by recording the amplification of a PCR product via a corresponding increase in the fluorescent signal associated with product formation during each cycle in the PCR. Quantification of gene (or transcript) numbers is determined during the exponential phase of the PCR amplification when the numbers of amplicons detected are directly proportional to the initial numbers of target sequences present within the environment. Quantification of the target gene during exponential amplification avoids problems that are associated with so-called ‘end-point’ PCR (in which amplicons are only analyzed after completion of the final PCR cycle) (Smith and Osborn, 2009).
In C. sativus, the development of the stigmas occurs concomitantly with transition of amyloplasts to chromoplasts and parallel with biosynthesis and accumulation of apocarotenoid which relates to expression levels of CsZCD and CsLYC gene (Bouvier et al., 2003; Rubio-Moraga et al., 2009). The pattern of accumulation of the apocarotenoid crocetin, picrocrocin and the different crocins in developed saffron stigmas was investigated by extracting stigmas from two selections (CITH-S-107 and PAM-S-116) with diverse crocin contents and stigma length (Tables 2 and 3). Biogenesis of crocetin glucosides and picrocrocin are initiated by zeaxanthin cleavage dioxygnase, which is coded by CsZCD gene (Rubio-Moraga et al., 2009). Although, carotenoid accumulation and composition during stigma development of C. sativus is highly regulated by the coordinated transcriptional activation of carotenoid biosynthetic genes (Castillo et al., 2005), the expression levels did not parallel the accumulation of the apocarotenoid compounds, suggesting that the formation of these compounds is controlled at a different level, such as carotenoid cleavage dioxygenase expression (Rubio-Moraga et al., 2008; Baghalian et al., 2010). Since, zeaxanthin cleavage being an important step in apocarotenoid accumulation, the authors choosed CsZCD gene expression to compare two divergent selections in saffron. The second gene studied in this experiment was CsLYC. Lycopene β-cyclase, the product of CsLYC gene, catalyzes the cyclization of lycopene to β-catrotene.
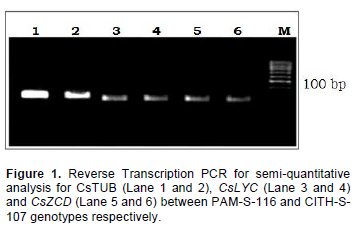
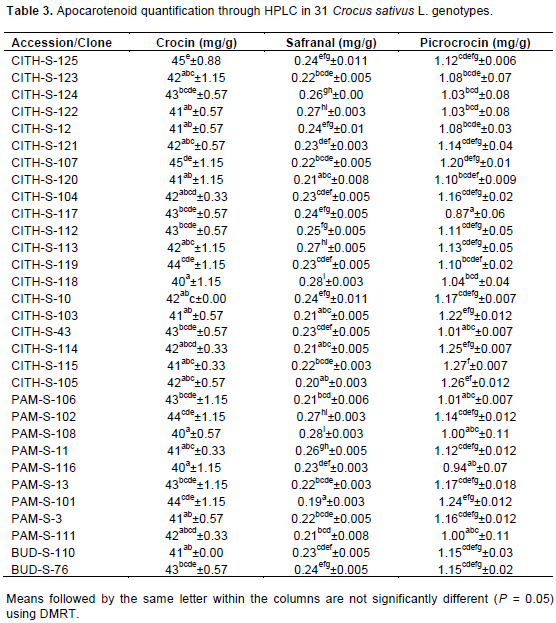
β-catrotene and zeaxanthin are the important precursors of saffron apocarotenoid in MVA pathway. Reverse transcription and real time PCR was performed with RNA purified from stigmas between two genotypes. Real Time PCR amplification of CsZCD, CsLYC and Tubulin genes between CITH-S-107 and PAM-S-116 at scarlet stages of stigma development is shown in Figure 2. PAM-S-116 showed 98 and 81% relative expression of gene CsZCD and CsLYC, respectively, to that of CITH-S-107 selection. CsZCD showed 3 fold higher expressions over CsTUB gene in both of the genotypes whereas CsLYC showed 0.7 and 0.3 fold expression over CsTUB in CITH-S-107 and PAM-S-116, respectively.
These results are in close agreement with the earlier reports of Mir et al. (2012a) who also reported 2.69 folds relative increase in CsZCD gene expression over tubulin gene expression in scarlet stage and 0.90 and 0.69 folds decrease in CsZCD gene expression over tubulin gene expression in orange and yellow stages, respectively. Increased expression of CsZCD gene with corresponding increase in apocarotenoid content during the development of stigma, suggests its regulatory role in apocarotenoid biosynthesis and stigma development in saffron (Mir et al., 2012a, b). Our results showed that variation in apocarotenoid content may be due to different expression levels of CsZCD and CsLYC genes between the genotypes.
The authors have not declared any conflict of interest.
Authors are thankful to Department of Biotechnology, University of Kashmir, Srinagar (J&K, India) and Central Institute of Temperate Horticulture (ICAR), Srinagar (J&K, India) for extended research and financial support.
REFERENCES
Baghalian K, Shabani Sheshtamand M, Jamshidi AH (2010). Genetic variation and heritability of agro-morphological and phytochemical traits in Iranian saffron (Crocus sativus L.) populations. Ind. Crops Prod. 31:401-406.
Crossref |
|
|
Bouvier F, Suire C, Mutterer J, Camara B (2003). Oxidative remodeling of chromoplast carotenoids: identification of the carotenoid dioxygenase CsCCD and CsZCD genes involved in Crocus secondary metabolite biogenesis. Plant Cell 15:47-62.
Crossref |
|
|
|
Caiola MG (2004). Saffron Reproductive Biology. Proc. Ist IS on Saffron Eds: J.-A. Fernández & F. Abdullaev. Acta Hortic. 650:25-37. |
|
|
Castillo R, Fernandez JA, Gomez-Gomez L (2005). Implications of Carotenoid Biosynthetic Genes in Apocarotenoid Formation during the Stigma Development of Crocus sativus and Its Closer Relatives. Plant Physiol. 139:674-689.
Crossref |
|
|
|
Freeman WM, Walker SJ, Vrana KE (1999). Quantitative RTPCR: pitfalls and potential. Biotechniques 26:112-125. |
|
|
|
Gomez-Gomez L, Rubio-Moraga A, Ahrazen O (2010). Understanding carotenoid metabolism in saffron stigmas: Unraveling aroma and color formation. Func. Plant Sci. Biotechnol. 4:56-63. |
|
|
Kanakis CD, Daferera DJ, Tarantilis PA, Polissiou MG (2004). Qualitative determination of volatile compounds and quantitative evaluation of safranal and 4-hydroxy-2, 6, 6-trimethyl-1-cyclohexene-1-carboxaldehyde (HTCC) in Greek saffron, J. Agric. Food Chem. 52:4515-4521.
Crossref |
|
|
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real- time quantitative PCR and the 2-∆∆CT Method. Methods, 25:402-408.
Crossref |
|
|
Mir JI, Ahmed N, Wafai AH, Qadri RA (2012a). Relative expression of CsZCD gene and apocarotenoid biosynthesis during stigma development in Crocus sativus L. Physiol. Mol. Biol. Plants 18(4):371-375.
Crossref |
|
|
|
Mir JI, Ahmed N, Wafai AH, Qadri RA (2012b). Variability in stigma length and apocarotenoid content in Crocus sativus L., selections of Kashmir. J. Spices Arom. Crops 21(2):169-173. |
|
|
Nehvi FA, Wani SA, Dar SA, Makhdoomi MI, Allie BA, Mir ZA (2007a). Biological interventions for enhancing saffron productivity in Kashmir. Acta Hortic. 739:25-31.
Crossref |
|
|
Nehvi FA, Wani SA, Dar SA, Makhdoomi MI, Allie BA, Mir ZA (2007b). New emerging trends on production technology of saffron. Acta Hortic. 739:375-381.
Crossref |
|
|
Pfander H, Schurtenberger H (1982). Biosynthesis of C20-carotenoids in Crocus sativus. Phytochemistry 21:1039-1042.
Crossref |
|
|
Rubio-Moraga A, Rambla JL, Ahrazem O, Granell A, Gomez-Gomez L (2009). Metabolite and target transcript analyses during Crocus sativus stigma development. Phytochemistry 70:1009-1016.
Crossref |
|
|
Rubio-Moraga A, Rambla JL, Santoella M, Gomez MD, Orzaez D, Granell A, Gomez-Gomez L (2008). Cytosolic and plastoglobule targeted caroteniod dioxygenase from Crocus sativus are both involved in β-ionone release. J. Biol. Chem. 283:24816-24825.
Crossref |
|
|
Sanchez AM, Winterhalter P (2013). Carotenoid Cleavage Products in Saffron (Crocus sativus L.). ACS Symp. Series 1134:45-63.
Crossref |
|
|
Smith CJ, Osborn AM (2009). Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol. Ecol. 67:6-20.
Crossref |