Molecular markers have proven to be powerful tools in research related with diversity, variability, and improvement of economically important tropical crops. This study analyzed eight physiological and morphological fruit characters of economic interest in the cultivated Mexican guava (Psidium guajava L.), and assessed the suitability of two sequence specific amplified polymorphism (SSAP) and simple sequence repeat (SSR) markers developed for their use in early selection of individual plants with given fruit characteristics. Principal component analysis (PCA) explained 79% of the morphological variability observed among accessions. S-SAP was more informative than AFLP for studies of variability and diversity in guava, the former marker showing higher percentage of polymorphism (90%) and more intraspecific variability (0.58). It was analyzed by cluster analysis using the unweighted pair group with arithmetic means (UPGMA) method the relationships between accessions from nine guava varieties. S-SAP dendrograms clustered varieties in better agreement with pulp color and fruit shape, suggesting a possible association of the S-SAP marker with quantitative trait loci (QTL) related to fruit physiological and morphological fruit characters. According to results, the microsatellites mPgCIR131, mPgCIR136 and mPgCIR161 might also be linked to QTL related to internal and external pulp thickness, pulp color, and soluble solids, indicating that the SSR markers developed are appropriate for their use in early selection of guava individuals having specific fruit features, therefore being suitable for molecular marker assisted selection (MAS) of the crop.
The guava (Psidium guajava L.) is a species of fruit tree belonging to the family Myrtaceae that is distributed in tropical and subtropical regions, mostly in Southeast Asia, Mexico, and Central and South America (Biffin et al., 2010). The trees and fruits of the species are worldwide known for their ecological and economic relevance (Grattapaglia et al., 2012; Woodrow et al., 2012), and in many countries guava fruits are highly valued, in some of them even being a staple food (Liu and Yang, 2011).
Some authors have proposed guava fruits are potentially nutraceutical due to their high contents of vitamins, minerals, and polyphenolic antioxidants (Hassimotto et al., 2005; Ho et al., 2012). Other parts of the guava plant have been used to treat diabetes, caries, wounds, diarrhea, inflammation, and hypertension (Gutiérrez et al., 2008), and have been reported as having anti-plasmodial, anti-inflammatory, hepatoprotective, anticancer, and antioxidant activities (Salib and Michael, 2004; Ojewole, 2006; Roy et al., 2006; Flores et al., 2015). The wide variety of applications and ecological importance of the species there is constant progress of numerous research efforts for improving its agronomical characteristics.
Conventional breeding methods to improve woody species as guava are limited (Rai et al., 2010; Liu and Yang, 2011). Selection of elite plants through the observation of phenotypic characters associated with traits of commercial importance continues to be favored among the methods for improvement of fruit tree crop production and the approach has in some cases proved to be effective, but breeding methods based on elite plant selection are also known to be extremely time consuming on average. In addition, the variation of the phenotypic characteristics in guava plantations has in many occasions proven to be mainly related to environmental factors, therefore being difficult to control (Srivastava and Narasimhan, 1967; Thaipong and Boonprakob, 2005).
In that context, molecular approaches have been not only useful for characterizing the genetic diversity among different guava cultivars, but also for identifying genes of commercial interest in the species. Among the most widely used molecular marker systems applied to studying the variability and genetic diversity of species in the genus Psidium are amplified fragment length polymorphism (AFLP), random amplified polymorphic DNA (RAPD), restriction fragment length polymorphism (RFLP), and simple sequence repeats (SSRs) (Ferreira et al., 1994; Risterucci et al., 2005; Chen et al., 2007; Hernández-Delgado et al., 2007; Krishna and Singh, 2007). Data obtained from AFLP, SSR, and RAPD markers have revealed an ample genetic variability in accessions of guava from Mexico and other Latin American countries (Domínguez-Álvarez et al., 2005; Risterucci et al., 2005; Aranguren et al., 2010; Padilla-Ramírez and González-Gaona, 2010; Valdés-Infante et al., 2010). However, the perspective remains open for development of molecular markers for detecting more polymorphisms and variability among Psidium guava accessions. The sequence-specific amplified polymorphism (S-SAP) marker approach developed by Woodrow et al. (2012) in the myrtle can provide molecular markers adequate for genetic variation and breeding studies of guava accessions.
In addition, recent molecular marker assisted selection (MAS) studies have indicated that SSR markers might be associated with specific quantitative trait loci (QTL) and this approach was shown to be highly efficient for selection of elite plants with characteristics of commercial importance (de Oliveira et al., 2012; Nimisha et al., 2013). Additionally, application based on MAS using SSR primers for selection of cultivars with certain phenotypic characteristics has been described in commercially important crops including among other, pepper (Minamiyama et al., 2007), rice (Fu et al., 2010), and apple (Kenis et al., 2008).
About 50 QTL for fruit characters and nearly 150 primers related to SSR have been described in guava that could be used for selection of elite plants with fruit characteristics of commercially interest (Risterucci et al., 2005; Ritter et al., 2010; Valdés-Infante, 2012). Because S-SAP markers could be more informative for studies of variability in guava than previously used markers or markers related to SSR primers, they could be applied for selection of elite plants. In this study LTR retrotransposons was identified by means of molecular analysis of accessions of P. guajava from Mexico and designed S-SAP markers for the species. These markers were evaluated and the results compared with previously developed AFLP markers for guava. The application of primers associated with simple sequence repeats (SRR) was also evaluated for selection of plants with fruit characteristics of commercial importance in populations of Mexican guava. The present research demonstrated the importance of the application of S-SAP and SRR markers for studies of variability and future selection in guava populations of productive characters, such as fruit features including content of vitamin C, pulp color, number of seeds, soluble solid content, and internal and external pulp thickness.
Plant materials
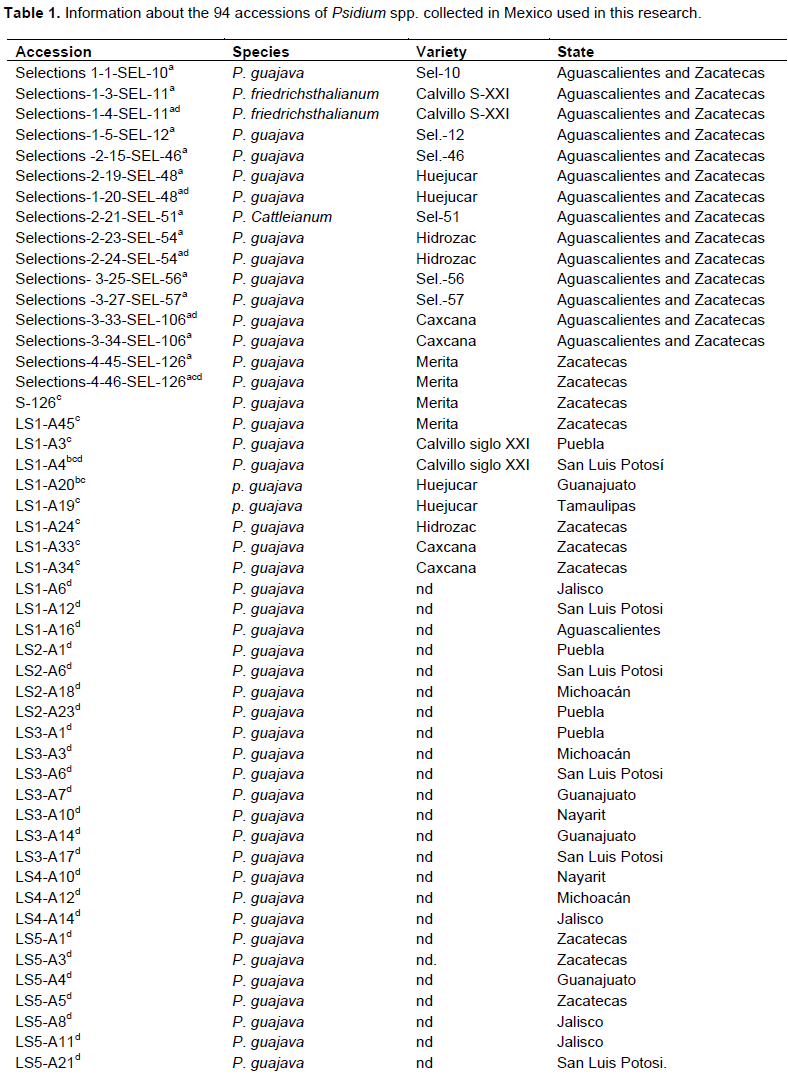
The present work used 91 accessions of guava collected from guava Germplasm Bank of the National Institute for Forestry, Agriculture and Livestock Research (INIFAP in its Spanish acronym), including five varieties (Calvillo siglo XXI, Caxcana, Huejucar, Hidrozac, and Merita) and three species (Psidium guajava, Psidium friedrichsthalianum and Psidium cattleianum). Nineteen accessions were used to assess variability through the SSAP marker and 70 accessions were used to validate the application of the use of SSR primers associated to external and internal pulp thickness (cm), pulp color, number of seeds, and soluble solid content (Table 1). Hernández-Delgado et al. (2007) described morphological characteristics of some of these accessions.
Description of trait of commercial importance in guava accessions
Seventy guava accessions were morphologically characterized (Table 1). The specific characteristics that were subjected to descriptive statistical analysis were: External and internal pulp thickness (cm), pulp color, number of seeds, and soluble solid content (°Bx). Degrees Brix were measured using an ATAGO model N-1EBX refractometer (Mercado-Silva et al., 1988) and the values of other features of interest were determined according to the procedures described by Hernández-Delgado et al. (2007).
Using the statistical package SAS version 9.0, a principal component analysis (PCA) based on the correlation matrix was made on average values of the quantitative and qualitative features fruit weight (g), polar diameter, equatorial diameter, thickness of external pulp (cm), thickness internal pulp (cm), soluble solids (°Bx), number of seeds, and pulp recorded from 70 accessions of Mexican guava from the states of Aguascalientes (3), Colima (3), Estado de Mexico (3), Guanajuato (5), Jalisco (8), Michoacan (5), Nayarit (10), Puebla (3) San Luis Potosi (8), Tabasco (2), Tamaulipas (1), and Zacatecas (19).
Development of S-SAP markers in guava
Long terminal repeat (LTR) retrotransposons of guava were isolated with the following procedure. DNA was extracted from young leafs from 16 guava accessions using the ChargeSwich kit (Invitrogen USA). Based on the protocol of Pearce et al. (1999), the extracted DNA was pooled (Table 1). The final concentration of the mixed sample was 1 mg of DNA in a final volume of 20 µL. The pooled DNA was digested with 1 U of the enzyme MseI at 37°C for 20 min. Afterwards, 50 pmol of MSE I adapters (5’-GACGATGAGTCCTGA G-3’ and 5’-TACTCAGGACTCAT-3’) were added to the digestion product, and ligated with 1 U of T4 ligase (New England Biolabs, USA) at 18°C during 16 h. Subsequently, 5 µL of the digested and linked DNA were used as a template for polymerase chain reaction (PCR) amplification with the RNase H1 (5’-MGNACNAARCAYATHGA-3’) and the MSEI (5’-GATGAGTCCTGAGTAANNN-) primers. Amplification conditions were as follows: One denaturing step at 94°C for 3 min, 30 cycles with one denaturing step at 94°C for 1 min, annealing at 45°C for 2 min, extension at 72°C for 2 min, and final elongation at 72°C for 10 min. The PCR products were separated by electrophoresis in 0.8% agarose gels and fragments between 400 and 2000 bp were selected, purified with the Wizard SV Gel and PCR clean up System (Promega, USA), and used in a second PCR with the RNase H2 (5-GCNGAYATNYTNACNAA-3’) primer and the previously described MSEI adapters (20 pmol.mL-1). The amplified fragments were cloned in the vector PGEM (Promega, USA) and transformed in competent E. coli cells. Twenty transformants that tested positive to containing the amplified fragments were selected and sent to MACROGEN Seoul, Korea, for sequencing.
Specific primers were designed for S-SAP in guava using the sequences of retroelements. Using BLASTn (http://blast.ncbi.nlm.nih.gov/), 5 sequences from the obtained PCR products were identified that aligned with ty1/Copia retroelements deposited in the GenBank (www.ncbi.nlm.nih.gov; Figure 2). Polypurine tract and LTR sequences were identified from ty1/Copia retroelement sequences from guava, from which five oligonucleotides were designed. The software packages Mega v. 4.1 (Tamura et al., 2013) and Oligo analyzer (http://www.idtdna.com/analyzer/applications/oligoanalyzer/Default.aspx) were used for sequence analysis.
Screening for the better set of primers for development of AFLP and S-SAP molecular markers
For selecting the better set of primers developed for the AFLP and S-SAP methods, five accessions belonging to different varieties were used: Calvillo Siglo XXI (LS1-A4), Caxcana (LS1-A33), Huejucar (LS1-A20), Hidrozac (LS1-A24), and Merita (LS1-A45) (Table 1). The ADN from these accessions of guava was extracted with the ChargeSwich kit (Invitrogen USA).
AFLP analysis
The AFLP method was performed following Tamayo-Ordóñez et al. (2012). Approximately 100 ng of genomic DNA were subjected to double-digestion by EcoRI and MseI endonucleases at 37°C for 1 h, and incubated at 65°C for 15 min. The DNA fragments were linked to EcoRI (5’-CTCGTAGACTGCGTACC-3’ and 5’-AATTGGTACGCAGTC-3’) and MseI (5’-GACGATGAGTCCTGAG-3’ and 5’-TACTCAGGACTCAT-3’) adapters at 15°C overnight. After pre-amplification with primers containing a single selective nucleotide (EcoRI primer 5’-GACTGCGTACCAATTCN-3’ and MseI primer 5’-GATGAGTCCTGAGTAAN-3’), a selective amplification by PCR was performed with four combinations of the EcoR1 and MseI primers. AFLP reactions with selective primers were performed with a touchdown PCR program for most primer sets with an initial denaturing step for 30s at 94°C, 30s for annealing at 65°C, and 1 min extension at 72°C. In each cycle, a decrease of 0.7°C in the annealing temperature was applied. Final annealing temperature was of 56°C, which was used for 24 cycles, with a final extension step at 72°C for 7 min. PCR products were electrophoresed in a CEQ 8800 sequencer (Perkin–Elmer Inc., Foster City, CA). The obtained electropherograms were analyzed using the software GeneMarker v.1.75 (Perkin-Elmer, Inc., Boston, MA).
S-SAP analysis
The S-SAP method was performed as described by Porceddu et al. (2002) and Vos et al. (1995). The S-SAP adapters and pre-amplification adapter primers used were the same as in Vos et al. (1995). A total of 20 primer pairs, obtained by the combination of the 5 retroelement primers and the 4 EcoRI primers were used for S-SAP (Table 2). In order to evaluate the reproducibility of the method, experiments were run by triplicate. The resulting bands were detected and analyzed by electrophoresis in a CEQ 8800 sequencer (Perkin-Elmer Inc., Foster City, CA). The combinations of primers used in the present study (Table 2) were performed with the touchdown PCR program previously described for AFLP.
Data analysis
AFLP and S-SAP bands were scored as 1 (present) or 0 (absent) in a binary matrix for each primer pair combination. Only reproducible and robust bands in each of the replications were considered as potential polymorphic markers. All calculations were performed using the NTSYS-pc 2.1 (Exeter Software Co., New York) software (Rohlf, 2000). In order to find out if the AFLP and S-SAP markers developed in this study could discriminate between accessions from different varieties of guava, nine accessions from five varieties were analyzed by the unweighted pair group with arithmetic means (UPGMA) method: Calvillo S-XXI (LS1-A3, LS1-A4), Huejucar (LS1-A19, LS1-A20), Hidrozac (LS1-A24), Caxcana (LS1-A33, LS1-A34), and Merita (LS1-A45, S126; Table 1). The genetic similarity between accessions was estimated by the simple matching genetic distance (SM) method calculated as: SM = m/n, where m is the number of matches and n is the total number of variables. Cluster analysis was performed from the similarity matrix by the UPGMA algorithm with the “FIND” option enabled to detect all possible trees.
The usefulness of AFLP and S-SAP markers for studying variability and polymorphisms was assessed using selected primers (Table 2) in 9 accessions of guava of which the variety was known, and which displayed phenotypic differences related to fruit features (Table 1).
Validation of the use of SSR primers as a tool for early selection in guava populations
The DNA from 70 guava accessions was extracted with the ChargeSwich (Invitrogen USA) kit (Table 1). Initially, a total of 18 SSR primers associated to 6 features of the fruit were chosen with the objective of selecting the best SSR primers for each fruit feature (Table 2). DNA from guava accessions having the highest and lowest values of each fruit characteristic, including internal and external thickness of the pulp, number of seed, soluble solid content, and pulp color was pooled and used as a template for amplification by PCR with the SSR primers specific to each described feature (Table 1). The values for each of the above-mentioned fruit features were subjected to analysis of frequencies in order to select the accessions of guava representing the minimum and maximum values of each feature to be used in the selection of SSR primers.
The PCR reaction conditions consisted of 2 ng of genomic DNA, PCR Buffer 1 X, 10 mM DNTPs, 10 pM each primer, 1.5 mM MgCl2, and 1U Taq polymerase in a final volume of 20 µl. Amplification conditions were as follows: One denaturing step at 94°C for 3 min, 35 cycles with one denaturing step at 94°C for 45 s, annealing at 55°C for 1 min, extension at 72°C for 30 s, and a final elongation at 72°C for 20 min. Polymorphic primers able to discriminate between contrasting fruit features were selected for each combination of oligonucleotides associated with each fruit characteristic, and monomorphic primers that amplified in both contrasting features were discarded.
Three primers were finally selected: mPgCIR161 as a marker of white pulp and lower soluble solid content, mPgCIR136 as a marker for individuals with small external pulp thickness, and mPgCIR131 as a marker of individuals with thicker internal pulp. Primers were labelled with the fluorophores FAM, NED, and JOE and DNA from 70 guava accessions was amplified. The amplified fragments were separated by capillary electrophoresis in an ABI PRISM 310 Genetic Analyzer (Applied Biosystems) and the visualization and classification by size of amplified fragments was performed by means of the software GeneMarker.
The results from GeneMarker were analyzed by means of a binary matrix with Microsoft Excel, indicating the absence and presence of fragments by the arbitrary values 0 and 1, respectively. The fragments that did not correspond to the expected size or having intensity peaks below 250 were eliminated from the matrix. Individuals were grouped according to allelic type (heterozygous or homozygous) and for each allelic type the percent efficiency of selection was calculated with the formula:
Characterization in individuals of guava of features of commercial interest
The descriptive statistics analyses of fruit characteristics in guava individuals indicated the following average values: 53.97±21.63 g of fruit weight, 10.76±1.52 °Bx of soluble solids, 184.28±69.95 seeds, 0.69±0.17 cm of external pulp thickness, and 3.69 ±0.48 cm of internal pulp thickness. Fruit weight and number of seeds proved to be the most variable morphological characteristics with coefficient of variation (CV) of 30%. The values of CV in the soluble solid content and in the external and internal pulp thicknesses varied from 10 to 24%.
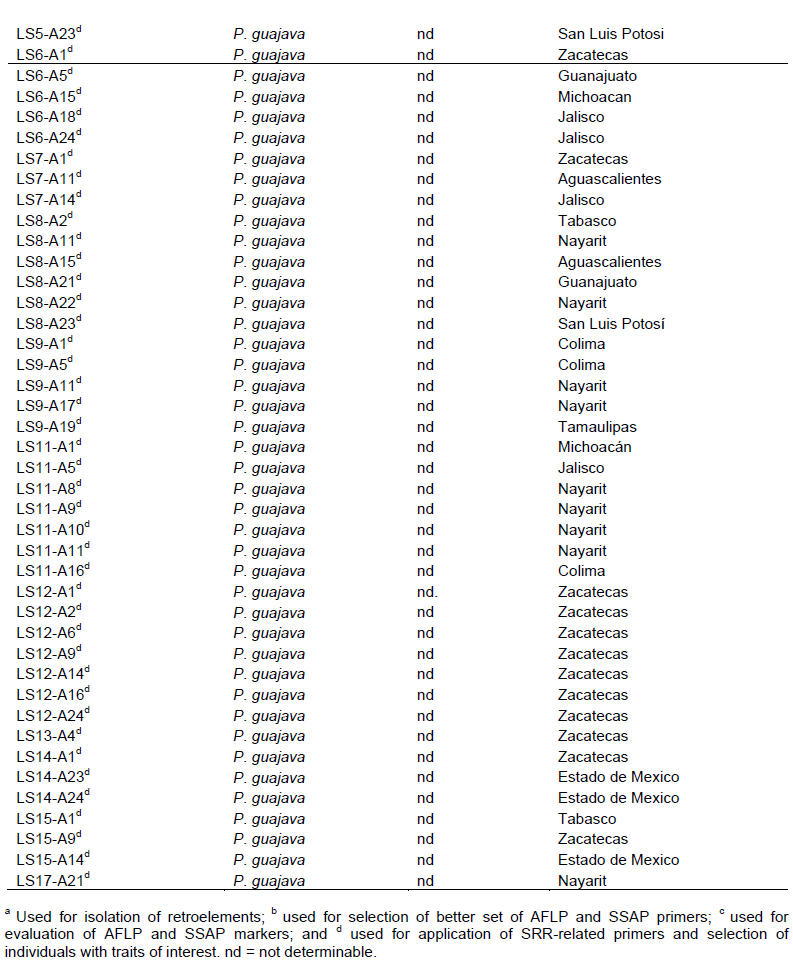
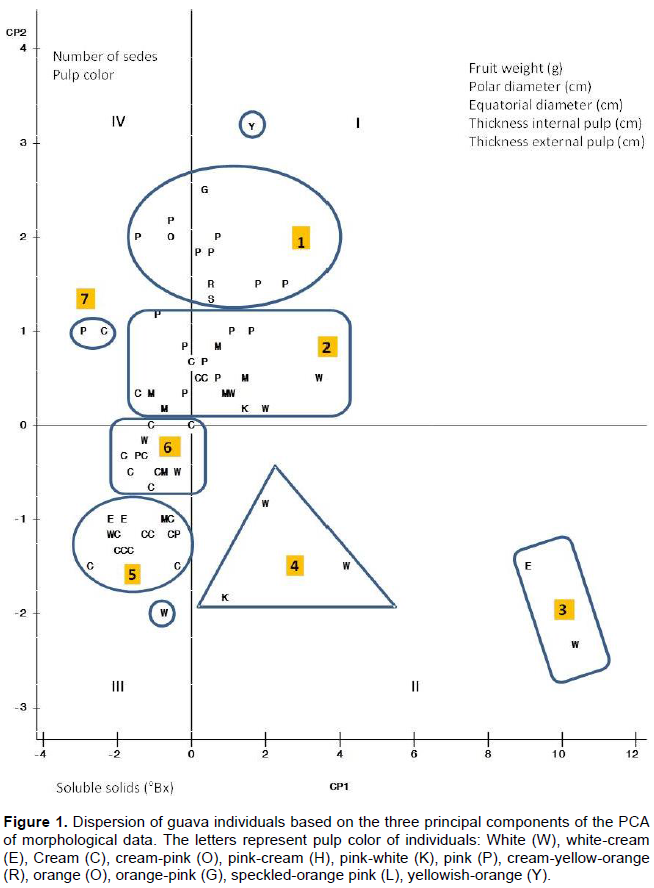
The results of PCA considering eight morphological characteristics (Table 1) indicated that the first two components (PC1 and PC2) explained 79% of the observed morphological variability (Table 4). According to the values of the eigenvectors, the characteristics with greater weight in PC1, all having positive values, were internal pulp thickness, external pulp thicknesses, and polar and equatorial fruit diameter, while in CP2 these characteristics were soluble solid content with negative value and, with positive value, seed number and pulp color (Table 2).
Figure 1 shows seven groups of guava accessions each group with the following average values for fruit characteristics: Group I with 11 accessions from Nayarit (7), Zacatecas (3), and State of Mexico (1), mostly with pink pulp, 64 g fruits, 4 cm thick inner pulp, 10 °Bx, and 284 seeds; Group II including two larger fruit accessions from Tabasco with white and creamy white pulp, 453 g fruits, 4 cm thick inner pulp, 10.7 °Bx, and 239 seeds; Group III with 16 accessions from Zacatecas (8), Jalisco (3), State of Mexico (2), Michoacán (1), Tamaulipas (1), and Guanajuato (2) with mostly with pink to creamy pink pulp, 72 g fruits, 7 cm thick inner pulp, 13.9 °Bx, and 402 seeds; Group IV with 3 accessions from Jalisco, Guanajuato, and Zacatecas with white and whitish pink pulp, 127 g fruits, 4.8 cm thick inner pulp, 13.5 °Bx, and 172 seeds; Group V with 15 accessions from Jalisco (4), Zacatecas (4), San Luis Potosí (2), Nayarit (2), Guanajuato (1), Michoacán (1), and Puebla (1) mostly with cream to whitish cream pulp, 37 g fruits, 14 °Bx, and 134 seeds; Group VI with 11 accessions from San Luis Potosi (3), Zacatecas (2), Guanajuato (2), Nayarit (1), Colima (1), Michoacan (1), and Puebla (1) mostly with cream pulp, 41 g fruits, 2.7 cm thick inner pulp, 8.6 °Bx, and 153 seeds; and Group VII including two accessions having the smallest and less sweet fruits from Puebla and Colima with pink to cream pulp, 20 g fruits, 2.7 cm thick inner pulp, 8.5 °Bx, and 123 seeds.
Identification of retroelements and selection of the better primers for development of AFLP and S-SAP molecular markers in guava
The bioinformatics analysis indicated presence of LTR-retroelements corresponding to the Ty1/Copia retrotransposon. From the obtained sequences it was possible to identify the ADIFTK RNase H2 motif, which is highly conserved in P. sativum, Diospyros kaki, and Vicia faba, among other species (Figure 2) (Pearce et al., 1999; Du et al., 2009). Also, ADIFTK RNaseH domains were shown to be present in myrtle (Myrtus communis L.), which belongs in the same plant family of P. sativum (Woodrow et al., 2012).
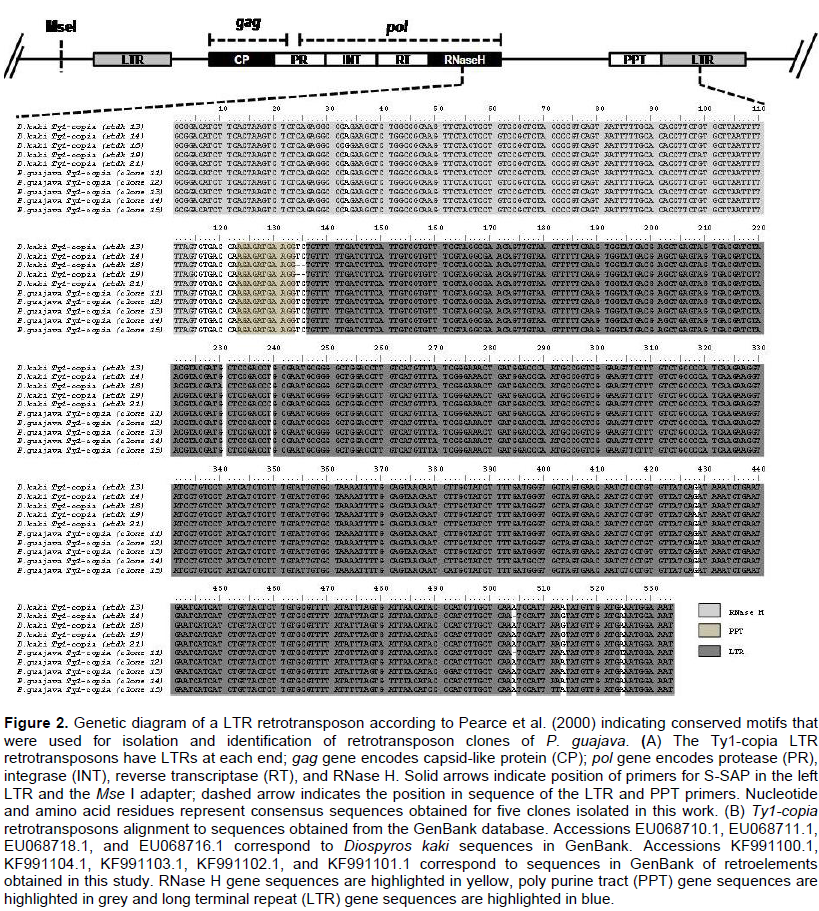
Polypurine tract (PPT) and long terminal repeat (LTR) sequences were identified in the isolated nucleotidic sequences. The Ty1/Copia retroelement sequences of studied guava accessions displayed a 90 to 100% similarity compared to the corresponding sequences of Diospyros kaki, with an average of 100% in nucleotide sequences. The Ty1/Copia retrotransposon sequences of P. guajava were submitted to the GenBank with the accessions numbers KF991100.1, KF991104.1, KF991103.1, KF991102.1, and KF991101.1.
From the latter sequences, the following 5 retroelement primers were designed: GUA1-5’-ATTGGGTCCATCAGTTTC-3’, GUA2-5’-ACACGAAATACGGCTACG-3’, GUA3-5’-CTGCGACTTCACCAAGCCAT-3’, GUA4-5’-CTTGAGGGGGAGTGTTGAG-3’, and GUA5-5’-AGGGAGGTCTAAACTGAGGAAA-3’.
AFLP showed a polymorphism of 78% (ACA/CTA) and 86% (AAG/CGC), and the tested combinations for S-SAP had a polymorphism of between 73 and 100% (Table 2). A total of 1,763 bands were analyzed for S-SAP, of which 1,560 (88.48%) were polymorphic. The primer pairs showing 100% polymorphism in S-SAP were GUA2/EcoR1(AAG), GUA3/EcoR1(AAC), GUA5/EcoR1(AAC), and GUA5/EcoR1(AAG) (Table 2). Unlike GUA2 and GUA3, primer GUA5 resulted in 97 to 100% polymorphism when being combined with any of the four EcoRI primers in the analyzed accessions.
Potential use of S-SAP markers for study of guava germplasm
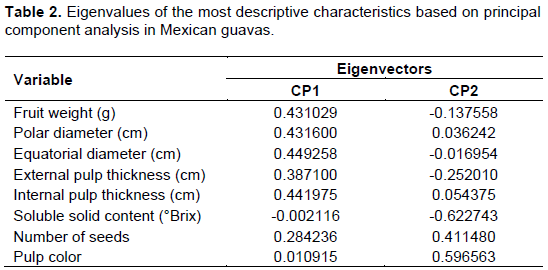
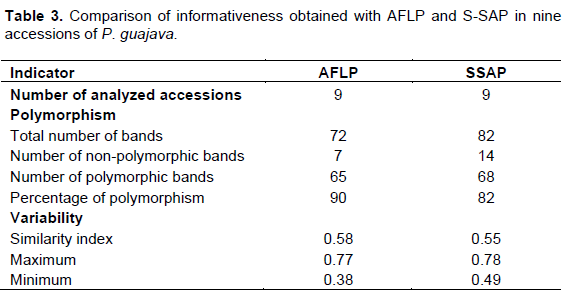
Table 3 contains the comparison of information provided by AFLP and S-SAP in the nine analyzed accessions for which the variety they belong to had been identified. Despite AFLP allowed for amplification of more bands than S-SAP (82 and 72, respectively), the number of polymorphic bands was similar for both compared methods (68 and 65, respectively). The percentage of polymorphism was higher when using the S-SAP method (90%) than when using the AFLP method (82%). The similarity indices between the nine analyzed accessions were 0.65 for AFLP and 0.58 for S-SAP.
The AFLP marker grouped eight of the nine analyzed accessions in a single cluster, while accession S-126 was grouped outside that cluster (Figure 3A).
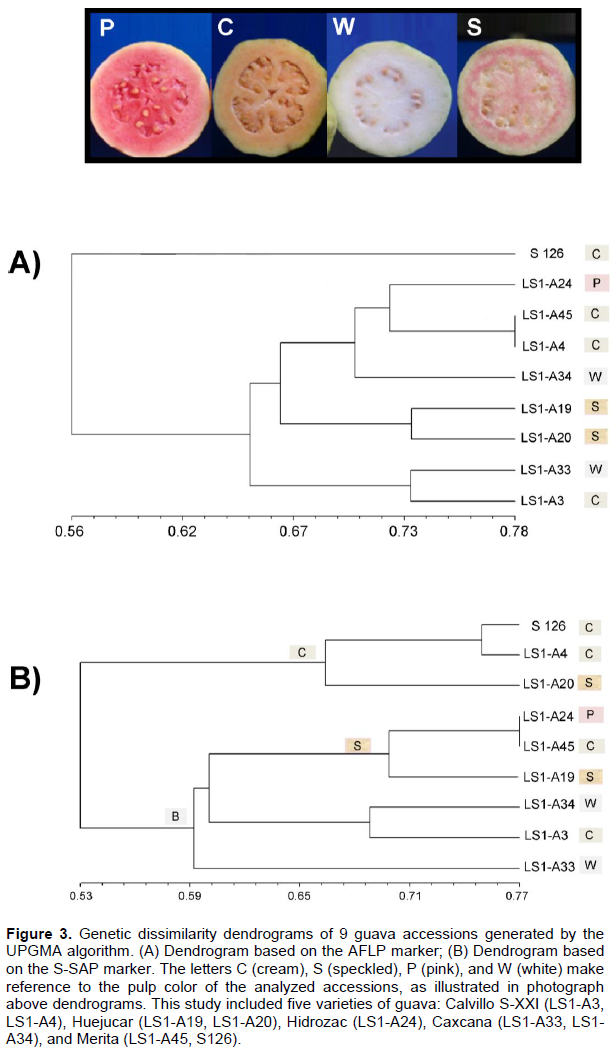
Contrastingly, the dendrogram obtained from the S-SAP marker showed two clusters (designated clusters I and II). Cluster I includes accessions S-126, LS1-A4, and LS1-A20, and cluster II groups accessions LS1-A24, LS1-A45, LS1-A19, LS1-A34, LS1-A3, and LS1-A33 (Figure 3B). No correlation was observed between the latter two clusters and the provenances of the accessions, but rather to morphological characteristics of the fruit such as pulp color and fruit shape, which are distinctive of each variety. Group I clustered together accessions with cream pulp and ovoid fruits. Group II with two subgroups clustered together varieties with white speckled pulp and truncate or semi-rounded fruits.
The results of the analysis of two sets of primers for developing S-SAP and AFLP markers (Table 3) showed that S-SAP produces 12% less bands than AFLP. However, despite the lower number of bands produced by S-SAP compared to AFLP, the former marker is more informative than the latter for studies of intraspecific variability and polymorphism in P. guajava. Previous studies indicated that the EcoRI/MseI (ACA/CTA and AAG/CGC) primer pair selected for AFLP showed a polymorphism of 84.2 and 92.4% in 68 accessions of P. guajava. However, the use of the latter set of primers in the present work in five varieties resulted in lower levels of polymorphisms than previously reported of 78 and 86%, respectively (Table 3), these values were nevertheless consider to be high enough for studying polymorphism and variability in guava. The level of polymorphism obtained for S-SAP (100%) is higher than that previously reported for AFLP (Hernández-Delgado et al., 2007). This means that S-SAP results in higher levels of polymorphism compared to AFLP. The observed indices of similarity between the nine analyzed accessions for AFLP and S-SAP indicate high intraspecific genetic variability. S-SAP showed lower similarity values than AFLP, which indicates that S-SAP allows for detecting more intraspecific variability in guava than AFLP. The results obtained indicate that S-SAP is more informative for studies of variability and polymorphism of P. guajava, as has been reported for other crops of economic relevance such as barley (Soleimani et al., 2005; Shan et al., 2012), grapevine (Stajner et al., 2015), holly (Levina et al., 2010), and citrus (Biswas et al., 2011), among others.
The high level of polymorphism obtained with marker S-SAP in comparison with AFLP was possibly due to the ubiquitous nature of retroelements (Flavell et al., 1992; Hirochika and Hirochika, 1993; Han, 2010), which suggests that the S-SAP marker could be well suited for studies of variability and diversity of Mexican guava germplasm. Otherwise, the resulting UPGMA dendrograms of nine accessions of guava showed discrepancies between the AFLP and the S-SAP approaches. AFLP formed a single cluster, while S-SAP formed two groups with the five analyzed varieties. S-SAP clustered varieties according to pulp color and fruit shape (Figure 3). Clustering of the cream, ovoid fruit varieties Calvillo Siglo XX-I and Merita in groups I and II could be indicative of intraspecific variability between these two varieties. Varieties Caxcana, Huejucar, and Hidrozac with white, speckled and pink pulp, respectively, clustered within group II. According to the distribution of varieties in the dendrogram, except for accession LS1-A3, most accessions with cream pulp tend to be located in the upper part of the dendrogram, accessions with speckled and pink pulp occupy the central part of the dendrogram, and accessions with white pulp were plotted at the bottom of the dendrogram.
PCA considering eight morphological characteristics indicated that the two first components explain 79% of the morphological variability observed among the accessions. Accessions grouped in five main groups in which pulp color showed not being determinant for discrimination of PCA groups. In contrast, clustering of S-SAP results suggests that the chosen primers hold great potential for their use in selection in guava of crops with specific pulp color, and could be used in early selection of elite material according to the agronomic characteristics of interest.
The grouping of accessions resulting from S-SAP according to pulp color and fruit shape may reflect an association of retroelements with a quantitative trait loci (QTL) conferring pulp color and fruit shape in guava. The association of retroelements with QTL related to fruit color has been described in economically important fruit crops such as peach (Falchi et al., 2013), grapevine (Kobayashi et al., 2004; Pelsy, 2010), and blood orange (Butelli et al., 2012). It has been suggested that high levels of activity of mobile elements (retroelements) can contribute to increment allelic diversity related to fruit color (Tao et al., 2005; Rico-Cabañas and Martinez-Izquierdo, 2007, De Felice et al., 2009; Butelli et al., 2012).
The color and shape of fruits are among the most popular commercial criteria for consumer acceptance of guava. However, low uniformity and quality in fruit have economic consequences for production and export of guava fruits. Despite the increasing interest in improving cultivars with desirable characteristics for consumers, little is known about the genetic factors that are related to the color and shape of fruit in guava, complicating the development of studies focused in marker assisted selection (MAS) of this crop (Collard and Mackill, 2008; Xu and Crouch, 2008). The possible association of retroelements with QTL related to pulp color and fruit shape in guava shown by the use of S-SAP marker could in the future be used for molecular mapping, marker assisted selection (MAS), and better understanding of gene function and gene regulation in guava, which would be of aid to the breeding of this crop species.
Simple sequence repeat (SSR) related to features of commercial importance
The evaluation of eighteen primer pairs associated to SSR related to six features of commercial importance (Table 3) indicated that only four primer pairs were polymorphic (discriminative between guava accessions with high and low values according to the feature analyzed), six were monomorphic (not discriminative between guava accessions with high and low values according to the feature analyzed discriminative), and six did not show amplification products.
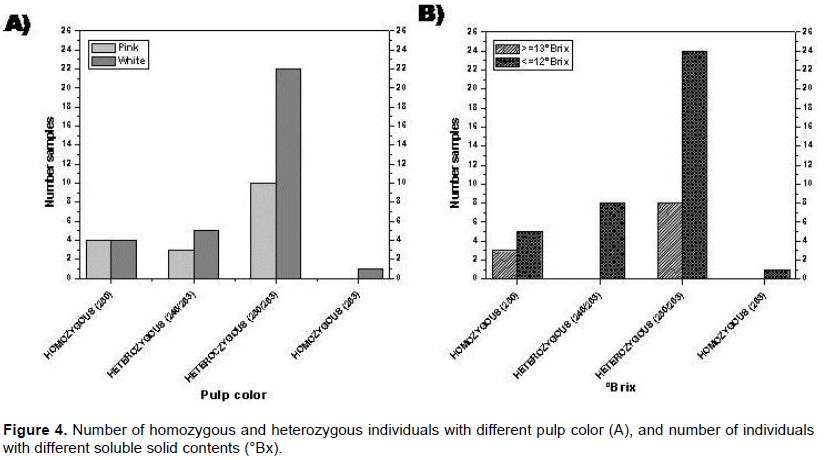
Three primers pairs (mPgCIR131, mPgCIR243, and mPgCIR208) were analyzed for evaluating the thickness of internal pulp: The mPgCIR131 pair was polymorphic for greater internal thickness; the mPgCIR243 pair showed no discrimination between accessions with higher or lower internal pulp thickness (was monomorphic); and the primer pair mPgCIR208 failed to amplify. Of the four primers pairs tested for association to external pulp thickness, three did not amplify (mPgCIR020, mPgCIR284, and mPgCIR243), only mPgCIR136 proving to be polymorphic for lower external pulp thickness (Figure 1A).
Regarding the two primers pairs (mPgCIR161 and mPgCIR254) associated with soluble solid content, mPgCIR161 only amplified in individuals having lower values of soluble solid content, while mPgCIR254 was found to be unable to discriminate between low and high soluble solid content values (Figure 1B). All of the three primer pairs (mPgCIR034, mPgCIR161, and mPgCIR208) evaluated for seed number were monomorphic and therefore not discriminative between low and high values.
The results obtained by the use of three SSR primer pairs associated with pulp color (mPgCIR079, mPgCIR137, and mPgCIR161) showed only mPgCIR161 was polymorphic for white and cream pulps, whereas primer pairs mPgCIR099137 and mPgCIR099 showed no amplification products (Figure 1C).
Finally, few guava accessions were characterized for vitamin C content and their DNA was not pooled. The primer pairs mPgCIR007 and mPgCIR094 showed being informative for selection of individuals with higher vitamin C content, but mPgCIR208 proved to be monomorphic for the feature (Figure 1D).
Based on the amplification patterns, the mPgCIR161 primer pair was finally selected as a marker of white pulp and lower soluble solids content, the mPgCIR136 primer pair was used for individuals with thinner external pulp, and the mPgCIR131 primer pair was selected as a marker of internal pulp thickness.
Improvement genetic in guava based in selection assisted by SSR markers
With the aim of validating the efficiency of the use of SRR primers in genetic improvement programs based on molecular markers, the different alleles amplified by each pair of primers assayed were analyzed. Alleles were classified in groups according to their size and homozygote (a single allele) and heterozygote (two alleles) types were discriminated. The assessed characteristics of commercial importance were internal and external pulp thicknesses, soluble solids, and pulp color.
Marker mPgCIR131 related to internal pulp thickness, showed presence of three amplification patterns in the analyzed guava accessions. Individuals from accessions LS3-A6, LS8-A2, and Selections-4-46-SEL-126 were homozygous with an allele sized at 148 bp, while individuals from other accessions were heterozygous with allele sizes of 141 and 148 bp. In the heterozygous alleles (141 and 148 bp), the calculated selection efficiency was 85%, indicating a value of internal pulp thickness of 3.84 mm. Homozygous alleles (148 bp) were unlinked to pulp thickness, being found in individuals with either thick or thin pulp.
In marker mPgCIR136 associated with external pulp thickness homozygous individuals with 110 or 120 bp alleles and heterozygous individuals with alleles of both sizes were observed. The selection efficiency calculated for the 110 bp allele was 100%, indicating a lower thickness of external pulp. The calculated average for each allelic type of external pulp thickness was 0.72, 1.3 and 0.90 mm for alleles 110, 110/120, and 120, respectively. Significant differences were also found between the homozygous individuals presenting the 110 or 120 bp alleles, suggesting the possibility that the 110 bp allele might be associated to thinner external pulp and that the 120 bp allele could be associated with thicker external pulp, heterozygous individuals having an intermediate value of external pulp thickness (1.3 mm).The marker mPgCIR 161 was associated with two features of agronomic interest, pulp color and soluble solid content. The results indicated homozygous individuals had allelic sizes of either 250 or 263 bp, and heterozygous individuals had alleles sized 246/263bp or 250/263bp. The selection efficiency calculated for the 250/263bp heterozygous allelic combination was 69% for selection of individuals with white pulp and 75% for selection of individuals with lower soluble solid content. Few homozygous or heterozygous individuals presented the alleles sized 250, 263, and 246/263bp, respectively, and they did not share pulp color or percent value of solid content, because of which the selection efficiency of these alleles could not be evaluated.
In the case of white pulp, the selection efficiency was 70%. All individuals homozygous for the 263 bp allele had white pulp, while one half of the homozygous individuals for the 250 bp allele had white pulp, and the remaining 50% of these individuals had pink pulp. Heterozygous individuals with both allelic combinations (246/263bp and 250/263bp) had a 1:2 a proportion of white pulp and pink pulp, suggesting that selection for white pulp might be based on the 263 bp allele (Figure 4A).
For the same mPgCIR-161 marker, the proportion of individuals having low values of soluble solid content appeared to be higher in presence of the 263 bp allele, which allows for concluding that this allele can be used for early selection of low soluble solid content and white pulp (Figure 4B).
The codominant nature of microsatellites facilitates the identification of homozygous and heterozygous individuals in early generations, which in turn allows for obtaining pure lines in the shortest time and discarding heterozygote lines from the beginning of the improvement process; an advantage over the traditional selection process (Zhou et al., 2003; Liu et al., 2007).
According to the information generated here, the SSR markers mPgCIR131, mPgCIR136, and mPgCIR161 could be linked to QTL related to internal and external pulp thickness, pulp color, and soluble solid contents. The variation observed in the selection efficiency values of each SSR allele associated with fruit characteristics was probably due to the different distances between the markers and the QTL (as closer the marker is to the QTL, the greater will be their association, which would increase efficiency) and to the observed variation in allele size for a same marker linked to a specified fruit property, which may be explained as the effect of the QTL on the feature. If the effect is less likely to be observed, it cannot be known if a given allele is responsible for a particular physiological characteristic (Maliepaard et al., 2001). These results indicated that the mPgCIR131 marker is linked to QTL related to internal pulp thickness and shows 46% of the variance of the character. This marker also presented selection efficiency of 85% and, according to data reported by Ritter et al. (2010), the distance to the QTL is 5.5 cM (centimorgans). It is well known that selection efficiency is closely related to the distance between the marker and the QTL. The recommended distance to QTL for successful selection based on linked markers has been reported to be of nearly 5 cM (Collard and Mackill, 2008).
In addition, the mPgCIR136 marker linked to the QTL associated with external pulp thickness explained 59% of the variance in the character and, also, it had a very high selection efficiency (100%), and was closer (5 cM) to the QTL related to this fruit characteristic (Risterucci et al., 2005; Ritter et al., 2010).
The mPgCIR161 marker related to pulp color and soluble solid content explained 81% of the variance and a selection efficiency of 75% of these characters of interest. The distance between mPgCIR161 and the QTL related to pulp color and soluble solid content was 13.2 cM (Ritter et al., 2010). According to the results obtained, it is possible that the use of the mPgCIR161 marker allows discriminating between individuals with pink and white pulp.
The low selection efficiency reported for the mPgCIR161 marker may have been influenced by genetic changes, such as deletions, insertions, transpositions, and DNA recombination, that may be affecting the association of the marker with the QTL specific to fruit color (Barton and Keightley, 2002; Juenger et al., 2005; Birchler and Veitia, 2010). Butelli et al. (2012) reported in other species that the movement of retroelements in the genome may be associated with change in coloration and that DNA recombination may be affecting the association of the marker with the QTL specific to this character; in some cases, recombination occurs between the QTL and the marker, theoretically, even if the distance between them is small (<5 cM). One centimorgan indicates that the probability of segregating separately is only 1% (Sharp et al., 2001; Thomas, 2003; Collard and Mackill, 2008; Rafalski, 2010).
In Mexico most guava crops were initially obtained through uncontrolled crosses, which increase the probability that genetic changes and recombination events resultant of hybridization of plants can affect the associations between markers and the QTL. By obtaining pure lines it is possible that the selection efficiency of marker associated to specific features will be increased (Anderson et al., 2001); which may be a future target for the improvement of guava.
There are few studies validating the use of markers for selection of fruit quantitative characteristics such as size and shape, softness, texture, and content of sugars. Other crops of commercial importance in which the SSR markers were shown to be associated to quantitative features of interest include papaya (Blas et al., 2011), potato (Li et al., 2013), and apple (Longhi et al., 2013), crops in which the use of these markers in improvement programs has been successful.
Furthermore, early selection would allow for obtaining pure lines with desired fruit characteristics in less time for satisfying the needs of producers, consumers, and the industry (Darwin-Robbins and Staub-Jack, 2009; Li et al., 2013). Currently desired guava phenotypes are white pulp, thick external and internal pulp, and high soluble solid content. The use of the SSR markers analyzed can be used for early selection of guava individuals with specific features and molecular marker assisted breeding can possibly improve this crop (Padmakar et al., 2015; Tuler et al., 2015).