ABSTRACT
Gua1 gene in Schizosaccharomyces pombe encodes inosine 5′-monophosphate dehydrogenase (IMPDH), which catalyzes the first step in de novo biosynthesis of guanosine monophosphate (GMP). Knock-out cassette was constructed with polymerase chain reaction (PCR)-based gene targeting technique for deletion of gua1 gene in S. pombe and this knock-out cassette was transformed to S.s pombe wild type (972h-). Knock-out cassette contains 653 and 285 bp sequences which are upstream and downstream of gua1 gene, respectively, in S. pombe genome and kanamycin resistance gene obtained from pFA6 plasmid. After transformation using lithium acetate method, knock-out cassette is aimed at replacing with the sequences of gua1 gene via homologous recombination. The transformant 972h- colonies which integrated knock-out cassette to the genome via homologous recombination are selected in YEA medium with antibiotic G418 after transformation and in this step, possible mutant colonies in gua1 gene were determined. Finally, colony PCR techniques were performed to control whether the deletion is made in the right place or not. The results show that the gua1 gene deleted strain was obtained.
Key words: Fission yeast, deletion, gua1 gene, knock-out cassette, inosine 5′-monophosphate dehydrogenase (IMPDH).
Inosine 5′-monophosphate dehydrogenase (IMPDH) is the key enzyme in de novo biosynthesis pathway of purine nucleotides. The role of IMPDH at metabolic branch point in de novo biosynthesis pathway of purine nucleotides makes IMPDH a target for drug discovery for immunosuppression (Ratcliffe, 2006), cancer (Chen and Pankiewicz, 2007; Oláh et al., 2006), antiviral (Nair and Shu, 2007) and antimicrobial chemotherapy (Shu and Nair, 2008). Schizosaccharomyces pombe is a free living unicellular fungus which has common features with higher eukaryotic organisms and characterized with properties of Ascomycetes (Wood et al., 2002). IMPDH is a product of gua1 gene in S. pombe and this gene is located near the centromeric region of chromosome II (Karaer et al., 2006).
Knock-out technology is carried out successfully in different organisms from single-celled eukaryotic organisms to mammalian including human cells (Kim et al., 2010; Shen et al., 2015; Rong and Golic, 2000). This technique is based on gene targeting and it modifies the genome of the living organism via homologous recombination. In gene targeting with homologous recombination technique, exogen DNA fragments (plasmid, cassette) produced in vitro are transformed into the cell in order to generate homologous recombination between target sequence and exogen DNA fragment which causes desired genetic change (Sawitzke et al., 2013; Gardner and Jaspersen, 2014).
Knock-out technology has made feasible research initiatives such as Saccharomyces Genome Deletion Project (http://www-sequence.stanford.edu/group/ yeast_deletion_ project/deletions3.html) which aimed to bring about a complete set of yeast deletion strains. Assigning specific functions to all open reading frames (ORFs) in one organism through phenotypic analysis of the mutants is the most common purpose of knock-out studies.
Korean BIONEER Corporation attempted to create a knock-out library for all S. pombe genes (http://pombe.bioneer.co.kr). The study aimed to obtain haploid mutant S. pombe with gua1 deletion, which does not take part in mutant collection of BIONEER. In this study, this cassette was similar to the modular kanMX6 cassette system described by Hentges et al. (2005) and Palabiyik et al. (2016). The lengths of homologous flanking sequences (60 to 80 bp) to the target gene were shorter in these studies, but our cassette had long sequences (653 to 285 bp) flanking to the gua1 gene. The gua1 gene deletion strain was obtained, although the strain is not viable for long time, as seen in other studies (Jiang et al., 2010).
Strains, media, growth conditions
S. pombe Linder str. Liquefaciens haploid wild type (972h-) and Escherichia coli DH5α strain containing pFA6-KanMX4 plasmid were used and these organisms were obtained from Istanbul University Department of Molecular Biology and Genetics Laboratory collection.
In this study, media which were described by Gutz et al. (1974) were used for production of S. pombe cells. S. pombe cells were produced in yeast extract with supplements (YES) media (supplemented with adenine, histidine, leucine, uracil, lysine and guanine at the concentration of 50 mg/L for each), yeast extract agar (YEA), minimal media agar (MMA). Luria Bertani (LB) media with ampicillin (50 mg/L) was used for E. coli containing pFA6-KanMX4 plasmid.
Isolation of pFA6-KanMX4 plasmid from E. coli DH5α strain and genomic DNA from S. pombe
E. coli DH5α strain containing pFA6-KanMX4 plasmid was grown overnight using LB media broth with ampicillin (50 mg/L) at 37°C and 150 rpm. Isolation of pFA6-KanMX4 plasmid was performed according to the manufacturer’s instructions (DSBIO Eco Friendly Plasmid Miniprep Kit).
972h- was grown in yeast extract agar plate 2 days at 30°C. Then, 1x 106 cells/mL were incubated overnight in yeast extract media at 30°C and 180 rpm. The isolation of genomic DNA from these cells was performed with DSBIO Quick Yeast Genomic DNA Extraction Kit according to the manufacturer’s instructions. Mechanical disruption was carried out with 0.3 g 0.45-0.55 mm glass beads in dismembrator (Sartorius Mikro-Dismembrator S) at 3000 rpm for 1 min and this step is repeated 5 times for each sample.
Production of knock-out cassette
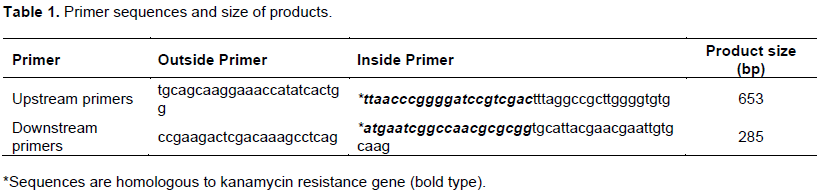
Primers which include both homologous sequences for downstream and upstream sequences of gua1 gene and short sequences that are homologous to kanamycin resistance gene are designed as shown at Table 1.
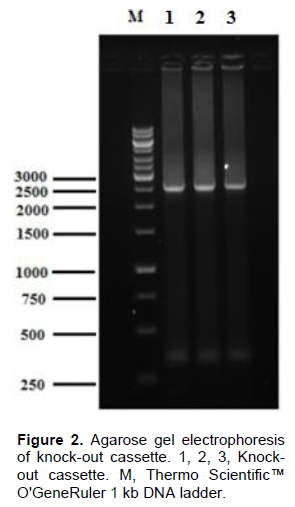
Knock-out cassette was obtained in two steps. In the first PCR, genomic DNA of S. pombe was used as a template and DNA fragments sized 652 and 285 bp were obtained. After purification, DNA fragments were used together for second PCR in which pFA6-KanMX4 plasmid was used as a template and outside primers. Knock-out cassette was observed by using agarose gel electrophoresis and gel extraction of cassette was carried out by Roche High Pure PCR Product Purification Kit.
Control of knock-out cassette with DNA sequence analysis
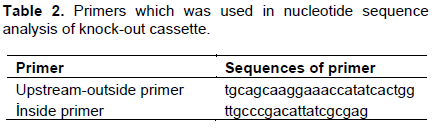
Upstream-outside primer of knock-out cassette and a reverse primer (inside primer) which complements kanamycin resistance gene (Table 2) were used in DNA sequence analysis. Sequence analysis was carried out with Sanger method by using 3500XL Genetic Analyzers (Applied Biosystems).
Transformation of knock-out cassette to S. pombe
Transformation of knock-out cassette to S. pombe 972h- wild type strain was carried out using lithium acetate method according to the method of Gregan et al. (2006). Solutions in transformation procedure (LiAc solution (0.1 M LiAc, 1X TE (pH 7.5)), LiAc-PEG solution (0,1 M LiAc, 40% PEG 3350, 1X TE (pH 7)) were sterilized by autoclave for 20 min at 121°C and 1.2 atm pressure.
Selection of transformant colonies
After the transformation of knock-out cassette to wild type S. pombe, for the detection of the transformant cells, these cells were inoculated into yeast extract agar plate with antibiotic G418 (200 mg/ml) (Sigma G418 disulfate salt) and incubated at 32°C for 5 days. Then resistant 972h- cells to antibiotic G418 were inoculated separately into selective media (YEA, YEA with G418, MMA and MMA with guanine) at the same time and incubated at 32°C for 5 days.
Control of transformant colonies
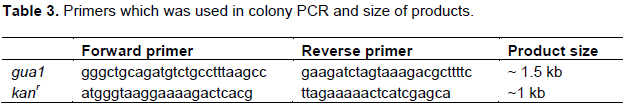
Colonies which grown with G418 were used as templates in PCR in order to determine the homologous recombination and colony PCR was separately performed with primers which are suitable for kanamycin resistance and gua1 genes (Table 3).
Production of knock-out cassette
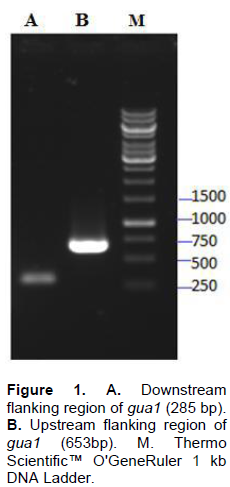
In the genome of S. pombe 653 and 285 bp homologous sequences which is located upstream and downstream flanking regions of gua1 gene respectively were amplified by two separate PCR (Figure 1). Knock-out cassette (2.5 kb) which includes kanamycin resistance gene was obtained at the second PCR (Figure 2).
Control of knock-out cassette with DNA sequence analysis
Results of DNA sequencing was confirmed with BLAST analysis in National Center for Biotechnology Information (NCBI). Sequencing results of knock-out cassette were compared with S. pombe 972h- genome and pFA6- KanMX4 plasmid. Outcomes of sequencing which carried out with upstream-outside forward primer were compared with genome of S. pombe 972h- and it was observed that this regions showed at the rate of 95% identification with downstream and upstream regions of gua1 gene.
Sequencing with inside reverse primer of kanamycin resistance gene was compared with pFA6-KanMX4 plasmid. According to the result of BLAST analysis, it was observed at the rate of 99% identification to kanamycin resistance gene and expectation value was found to be “0.0”.
Observation of null gua1 mutant of S. pombe
24 independent transformation experiments with the knock-out cassette yield, 3604 positive colonies which were obtained according to kanamycin resistance, was carried out. Subsequently, colonies were inoculated into YEA, MMA, YEA with G418 and MMA with guanine in order to confirm whether cassette was inserted to gua1 gene region or not. 56 colonies which could not live in media without guanine were detected after incubation for 5 days at 32°C.
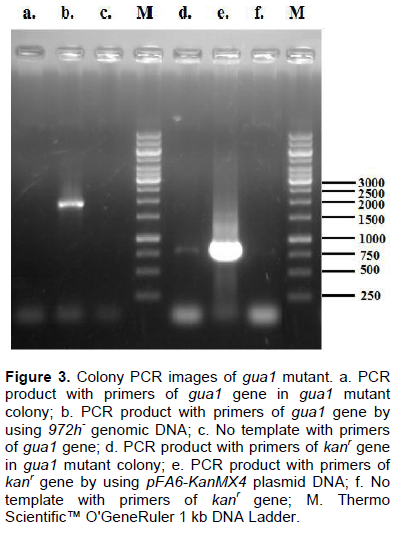
After colony PCR with these 56 transformant colonies and primers which are appropriate to gua1 and kanr gene, samples were analyzed on agarose gel electro-phoresis. In 55 colonies, it was observed that insertion of knock-out cassette into the location of gua1 gene was unsuccessful. Gua1 gene fragment does not exist in only one colony but a band with primers of kanr gene in the PCR of this colony (Figure 3).
Observation of gua1 deletion mutant cells using the phase-contrast microscope
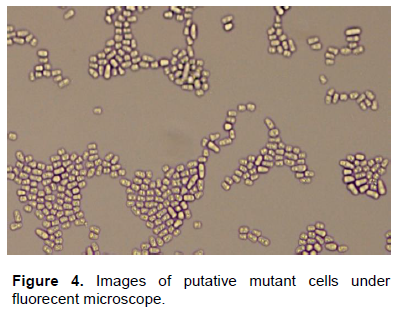
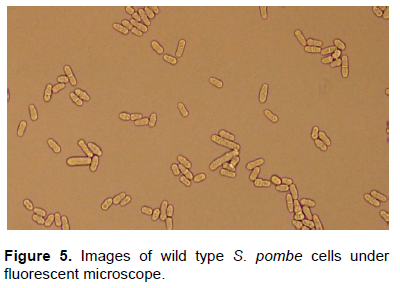
When gua1 deletion mutant cells (Figure 4) and wild type S. pombe cells (Figure 5) were investigated under the fluorescent microscope, it was seen that mutant cells were smaller than wild type and mutant cells tend to be aggregate.
Over the past 60 years, S. pombe has become an important model organism along with S. cerevisiae to study the molecular biology of eukaryotes (Hoffman et al., 2015). Yeasts help to improve comprehension of basic cellular processes and metabolic pathway in human. This organism makes easier molecular analysis of several genes related to diseases (Suter et al., 2006). S. pombe is a very convenient model organism for investigating specific cancer pathway, owing to its cell cycle and cell control point mechanisms (Wood et al., 2002).
Deletions of S. pombe’s ORFs were carried out at the rate of 98.4% (Kim et al., 2010; Spirek et al., 2010). Gua1 gene in which deletion was performed in this study was not yet deleted within the scope of the “S. pombe Genome Deletion Project”. Obtaining a viable gua1 gene deleted strain will make a contribution regarding the determination of the effect of IMPDH on the various cellular pathway such as cancer and immunosuppression.
In this study, the deletion of gua1 gene was carried out by creating knock-out cassette via homologous recombination according to Kim et al. (2010). Before transformation knock-out cassette, it was analyzed with sequencing.
Colonies which were phenotypically selected were confirmed by colony PCR. In the results of colony PCR, one single candidate transformant colony from 972h- showed the expected band profile on the agarose gel electrophoresis. PCR was performed in the candidate 972h- colony using primer set of kanr gene, the band of interest at the correct size was observed. However, no band was shown when PCR was carried out with same colony but using primer sets of gua1 gene. After the transformation, 56 colonies were reproduced at selective media and it was observed that only one colony was inserted knock-out cassette to the target region in genomes.
When these mutant cells were analyzed micro-scopically, it was observed that the cells were small and displayed aggregation similar to the results of Fernández-Álvarez et al. (2009). In this study, these mutant colonies were phenotypically more flattened than wild type colonies. Jiang et al. (2010) studied on human fungal pathogen Candida albicans and aimed at the deletion of gua1 gene in Candida albicans. They reported that gua1/gua1 diploid mutant colonies could not live longer than a week at 4°C. Similar to the results of Jiang et al. (2010), it is also observed that these mutant cells with gua1 deletion are not viable at 4°C for a long time. The authors passaged this mutant cells six times in every other day and inoculated at 30°C, then it was observed that the mutant cells can survive for six or seven generations. This study could be maintained with glycerol stocks of the single colony which was obtained after the transformation. Although, mutant cells with deletion of gua1 gene have no long life span, the deletions in their genome are permanent and after some optimization, this mutant strain could be used in other studies.
Decottignies et al. (2003) carried out a pilot gene deletion project in S. pombe to determine the number of essential genes. They reported that, obtaining systematic deletion of all S. pombe ORFs is more difficult than S. cerevisiae when similar studies are compared. The reason is that the homologous recombination efficiency of S. pombe is lower than S. cerevisiae (Kaur et al., 1997).
In addition, they reported that the genome of S. pombe includes some regions which contain inadequately transcribed genes and these genes have closed chromatin structure. The other possible reason for low efficiency may be the deletion cassette integrated into genome but kanamycin resistance gene was deficiently expressed by the reason of heterochromatin structure (Decottignies et al., 2003). The location of a gene in the centromeric region reduces the homologous recombination frequency. In this context, location of the gua1 gene makes the deletion to be more difficult than the other genes which are in euchromatic region. Finally, data presented here support the fact that the obtained colony is the first mutant strain of S. pombe with deletion of gua1 gene.
The authors declare that there is no conflict of interest.
REFERENCES
|
Chen L, Pankiewicz KW (2007). Recent development of IMP dehydrogenase inhibitors for the treatment of cancer. Curr. Opin. Drug Discov. Dev. 10(4):403-412.
|
|
|
|
Decottignies A, Sanchez-Perez I, Nurse P (2003). Schizosaccharomyces pombe essential genes: a pilot study. Genome Res. 13:399-406.
Crossref
|
|
|
|
|
Fernández-Álvarez A, Elías-Villalobos A, Ibeas JI (2009). The O-mannosyltransferase PMT4 is essential for normal appressorium formation and penetration in Ustilago maydis. Plant Cell 21:3397-3412.
Crossref
|
|
|
|
|
Gardner JM, Jaspersen SL (2014). Manupulating the yeast genome: deletion, mutation, and tagging by PCR. Methods Mol. Biol. 1205:45-78.
Crossref
|
|
|
|
|
Gregan J, Rabitsch PK, Rumpf C, Novatchkova M, Schleiffer A, Nasmyth K (2006). High-throughput knockout screen in fission yeast. Nat. Protoc. 1(5):2457-2464.
Crossref
|
|
|
|
|
Gutz H, Heslot H, Leupold U, Loprieno N (1974). Schizosaccharomyces
|
|
|
|
|
Hentges P, Van Driessche B, Tafforeau L, Vandenhaute J, Carr AM (2005). Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast 22(13):1013-1019.
Crossref
|
|
|
|
|
Hoffman CS, Wood V, Fantes PA (2015). An ancient yeast for young geneticists: a primer on the Schizosaccharomyces pombe model system. Genetics 201:403-423.
Crossref
|
|
|
|
|
Jiang L, Zhao J, Guo R, Li J, Yu L, Xu D (2010). Functional characterization and virulence study of ADE8 and GUA1 genes involved in the de novo purine biosynthesis in Candida albicans. Fems. Yeast Res. 10(2):199-208.
Crossref
|
|
|
|
|
Karaer S, Topal-Sarıkaya A, Arda N, Temizkan G (2006). The 3' terminal sequence of the inosine monophosphate dehydrogenase gene encodes an active domain in the yeast Schizosaccharomyces pombe. Genet. Mol. Biol. 29(3):551-557.
Crossref
|
|
|
|
|
Kaur R, Ingavale SS, Bachhawat AK (1997). PCR-mediated direct gene disruption in Schizosaccharomyces pombe. Nucleic Acids Res. 25:1080-1081.
Crossref
|
|
|
|
|
Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, Han S (2010). Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 28:617-623.
Crossref
|
|
|
|
|
Nair V, Shu, Q (2007). Inosine monophosphate dehydrogenase as a probe in antiviral drug discovery. Antivir. Chem. Chemother. 18(5):245-258.
Crossref
|
|
|
|
|
Oláh E, Kokeny S, Papp J, Bozsik A, Keszei M (2006). Modulation of cancer pathways by inhibitors of guanylate metabolism. Adv. Enzyme Regul. 46:176-190.
Crossref
|
|
|
|
|
Palabiyik B, Jafari Ghods F, Karaer Uzuner S (2016). Role of glucose repression in the oxidative stress response of Schizosaccharomyces pombe: analysis of transcript levels of fbp1, hxk2, sod1 and ctt1 genes in sty1, atf1 and pap1 knock-out mutants. Turk. J. Biol. 40:815-825.
Crossref
|
|
|
|
|
Ratcliffe AJ (2006). Inosine 5'-monophosphate dehydrogenase inhibitors for the treatment of autoimmune diseases. Curr. Opin. Drug Discov. Dev. 9(5):595-605.
|
|
|
|
|
Rong YS, Golic KG (2000). Gene Targeting by Homologous Recombination in Drosophila. Science 288:2013-2017.
Crossref
|
|
|
|
|
Sawitzke JA, Thomason LC, Bubunenko M, Li X, Costantino N, Court DL (2013). Recombineering: using drug cassettes to knock out genes in vivo. Methods Enzymol. 533:79-102.
Crossref
|
|
|
|
|
Shen B, Xiao J, Dai L, Huang Y, Mao Z, Lin R, et al. (2015). Development of high-efficiency gene knockout system for Pochonia chlamydosporia. Microbiol. Res. 170:18-26.
Crossref
|
|
|
|
|
Shu Q, Nair V (2008). Inosine monophosphate dehydrogenase (IMPDH) as a target in drug discovery. Med. Res. Rev. 28:219-232.
Crossref
|
|
|
|
|
Spirek M, Benko Z, Carnecka M, Rumpf C, Cipak L, Batova M, Marova I, Nam M, Kim DU, Park HO, Hayles J (2010). S. pombe genome deletion project: an update. Cell Cycle 9(12):2399-2402.
Crossref
|
|
|
|
|
Suter B, Auerbach D, Stagljar I (2006). Yeast-based functional genomics and proteomics technologies: the first 15 years and beyond. Biotechniques 40:625-644.
Crossref
|
|
|
|
|
Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D (2002). The genome sequence of Schizosaccharomyces pombe. Nature 415:871-880.
Crossref
|
|