ABSTRACT
Pseudomonas aeruginosa is a Gram negative aerobic rod shaped bacterium and is an opportunistic pathogen that usually causes nosocomial infection in immunocompromised patient with various infections and affects normal healthy human as well. P. aeruginosa is also an omnipresent pathogen that can be inhabited in soil, water, vegetable, human and animal. Metallo-β-lactamases (MBL) producing P. aeruginosa are known to be resistant to almost the entire anti-pseudomonas agent via mechanism of low outer membrane permeability, β-lactamases synthesis and the efflux systems. This study was conducted to detect the potential of metallo-β-lactamases producing P. aeruginosa presence in water samples from various parts of Malaysia. In this study, 52 water samples were collected from various parts of Malaysia. These P. aeruginosa isolates were processed to these phenotypic methods, Hodge test which is used to detect the carbapenemase production, Imipenem- EDTA combined disc test (CDT). Imipenem-EDTA double disc synergy test (DDST) was used to determine metallo-beta-lactamases producing P. aeruginosa. Among 52 various sources of water samples, 13 water samples had positive P. aeruginosa isolates. 6 out of 13 positive isolates have shown positive results in Hodge test, CDT and DDST. All metallo-β-lactamases producing isolates are multi-drug resistant. Among water samples from various parts of Malaysia, MBL producing P. aeruginosa are highly found in Selangor followed by KL, and this P. aeruginosa are detected from drain followed by river. Although some advantages and disadvantages exist among these phenotyping methods, still CDT and DDST are optimal to detect MBL.
Key words: Metallo-β-lactamases, multi-drug resistance, phenotyping method, Pseudomonas aeruginosa.
Pseudomonas aeruginosa is an opportunistic pathogen that usually causes nosocomial infection in immuno-compromised patient with severe infections, such as cystic fibrosis, cancer, burn, urinary tract infection. They can also affect normal healthy human, which might cause dermatitis conjunctivitis, otitis externa and gastrointestinal infection (Hallin et al., 2012; Adesoji et al., 2015). P. aeruginosa is a highly adaptable bacterium, can survive in a wide range of environment and change its properties in response to changes in the environment (Lambert, 2002). The outbreaks of P. aeruginosa that are associated with different water sources such as drains, lakes and wastewater had been reported by Quick et al. (2014).
Antibiotic-resistant bacterial can be found in water sources from hospital, industrial or domestic wastes into watercourses (Chartier et al., 2014). It is due to contribution of the un-metabolized antibiotics in the water, which is excreted from humans. It may be a potential risk causing problem to human health (Email et al., 2010). Hospital wastewaters contain antimicrobial resistant bacterial with a level of at least a factor of 2 to 10 times higher than in domestic wastewater. Antimicrobial resistant bacterial gene transfer is great at high cell densities and under high antibiotic concentrations (Chartier et al., 2014). The effluents of the industries also act as one of influence on the pollution of the water bodies; these effluents can change the physical, chemical and biological nature of the water body (Hussain and Rao, 2013).
The characteristic features of the climate in Malaysia are uniform temperature, high humidity and ample rainfall. Malaysia mainly has 2 seasons, which are southwest monsoon season and northeast monsoon season (General Climate of Malaysia, 2016). November, December and January are the months with maximum rainfall, in the east coast while June and July are the driest months in most districts. Over the rest of the Peninsula with the exception of the southwest coastal area, the monthly rainfall pattern shows two periods of maximum rainfall separated by two periods of minimum rainfall (Chen et al., 2013; General Climate of Malaysia, 2016). Altering climatic conditions can change the volume and quality of water availability in both time and space, influencing the water usage practices. For an example, intensity of rainfall, or the period of time without rain will affect the quality of water in rivers and lakes through alteration in the timing and volume and temperature. Besides that, floods also have a probability of spreading diseases by over following the open sewage or inadequate sewage infrastructure (Corcoran et al., 2010).
Phenotyping methods are based on the ability of metal chelators, such as EDTA and thiol-based compounds, inhibit the activity of MBL (Manoharan et al., 2010). Double Disk Synergy Test (DDST) and Combined Disc Test (CDT) are most commonly used to detect the MBL producing P. aeruginosa then Hodge test. Modified Hodge test detects only carbapenemase activity, which prevents the use of EDTA and therefore, does not confirm the metal dependence of the carbapenemase (Kali et al., 2013). Modified Hodge test, DDST and the CDT are easy, reliable, simple to perform and cheaper (Qu et al., 2009). The CDT is the most sensitive techniques for detecting MBL compared to DDST (Biradar and Roopa, 2015). DDST is used to distinguish MBL producing gram negative bacilli from MBL non producing gram negative bacilli. For those MBL producing P. aeruginosa isolates positive by DDST, are also positive by CDT for MBL production (Bhalerao et al., 2010).
Most of the studies were concentrated on the isolation of β- lactamase gene from clinical samples (Kumar et al., 2012; Upadhyay and Joshi, 2015). Therefore, the aim of present study is to detect the potentiality of MBL producing P. aeruginosa in water samples from various parts of Malaysia. The objective of this study is to determine the presence of P. aeruginosa from water source, to evaluate the MBL producing P. aeruginosa by using modified Hodge test, double disk synergy test (DDST) and combined disk test (CD).
Bacteria isolation
Fifty-two water samples of different water source were collected from various parts of Malaysia. Each water sample was then diluted by using serial dilution method, 10-1 to 10-5. The diluted sample was inoculated on the Nutrient agar (NA), Cetrimide Nalidixic Acid agar (CA), Blood agar (BA) and MacConkey agar (MAC). The inoculated agar was incubated at 37ËšC for 16 to 24 h Ce´ line (Slekovec et al., 2012; Pellegrini et al., 2009).
Characterization and identification
The colonies on the Nutrient agar, Cetrimide Nalidixic Acid agar, Blood agar and MacConkey agar were observed for morphological characteristics and were processed for various identification tests (Nasreen et al., 2015) such as, gram staining to distinguish gram positive bacteria from gram negative bacteria; oxidase test to differentiate pseudomonas from enterobacteriaceae, and biochemical tests (Indole, Methyl red, Voges-Proskauer, citrate, urease and triple sugar iron) to identify the species of bacteria (James and Natalie, 2007).
Antibiotic susceptibility testing
Antibiotic susceptibility test was carried out on P. aeruginosa isolates by using the disc diffusion and Kirby-Bauer methods, in accordance to the Clinical and Laboratory Standards Institute (CLSI, 2011) guidelines (Bashir et al., 2011).The identical colonies in bacterial culture from nutrient agar were inoculated into nutrient broth and were incubated at 37ËšC for 3 to 4 h. The inoculum density was adjusted to a MacFarland 0.5 standard and it was inoculated on the Mueller-Hinton agar plate using a lawn culture technique. The inoculated plate was allowed to dry (Nasreen et al., 2015).
The following antibiotics were placed on the inoculated agar by disc diffusion method, Ciprofloxacin (5 µg), Gentamicin (10 µg), Amikacin (30 µg), Co-trimoxazole (25 µg), Tetracyline (30 µg), and Cefuroxime (30 µg). The plates were incubated at 37°C for 16 to 18 h. The zone of inhibition around the disk were measured and categorized into susceptible, intermediate and resistant.
Hodge test
The Hodge test was performed by preparing a 0.5 McFarland dilution of E.coli ATCC 25922 in 5 ml of nutrient broth. The diluted E. coli indicator organism was streaked on the Mueller Hinton agar using lawn culture technique and allowed to dry for 3 to 5 min. The test organism in a straight line from edge of disk to the edge of the plate and placed a carbapenem disc. The carbapenem disc is Imipenem (10 µg) or Meropenem (10 µg) placed at the center of the plate. The plates were incubated at 37°C for 16 to 24 h (Chaudhari et al., 2011; Georgios et al., 2014).
Imipenem-EDTA double- disc synergy test (DDST)
DDST was performed in accordance to the CLSI recommendations for the disk diffusion method. 0.5 McFarland of test organism was streaked on the Mueller Hinton agar by using sterile cotton swab to get a lawn culture. Placed imipenem (10 ug) disc in 20 mm center to center from a blank disc contained 10 µl of 0.5 M EDTA (750 ug) or EDTA disc (750 ug). The plate was incubated at 37°C for 16 to 18 h (Bhalerao et al., 2010; Chaudhari et al., 2011; Kali et al., 2013).
Imipenem-EDTA combined disc test (CDT)
0.5 McFarland of test organism was streaked on Mueller Hinton agar by using sterile cotton swab to get a lawn culture and was allowed to dry for 3 to 5 min. Placed 2 imipenem (10 ug) disc on the agar at distance of 25 mm. 10 µl of EDTA solution was added to one of them to obtain desire concentration of 750 µg. Then the plates were incubated at 37°C for 16 to 18 h (Arunagiri et al., 2012; Kali et al., 2013).
Bacteria isolation and identification
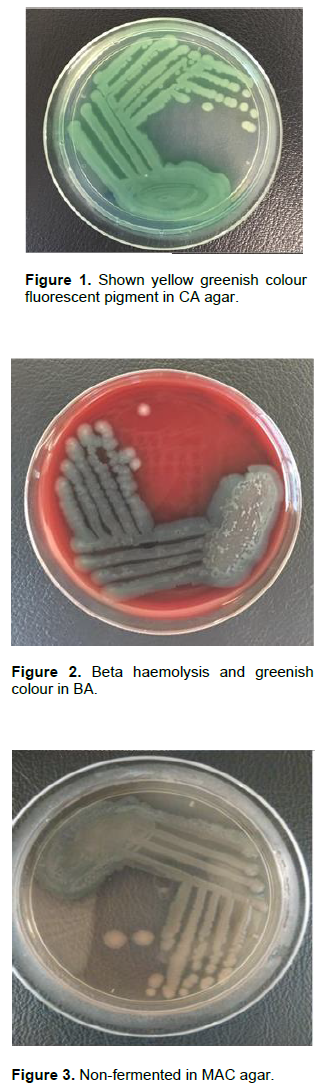
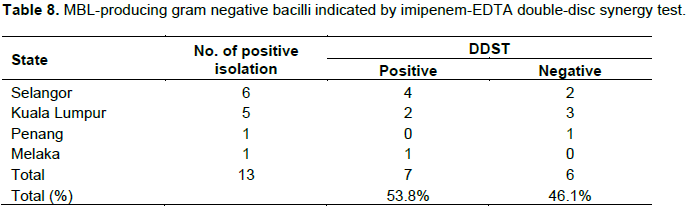
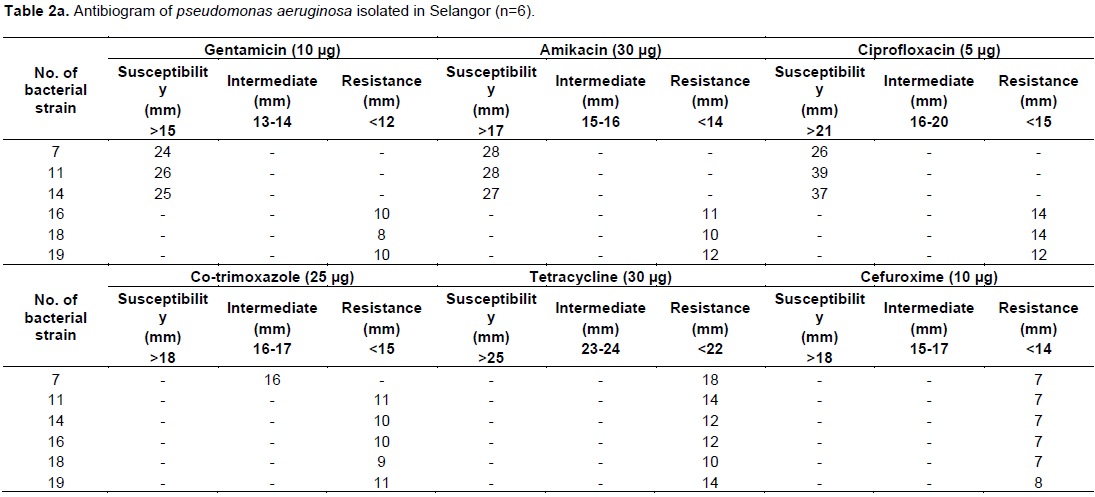
In this study, among 52 water samples, 20 water samples from Selangor, 15 water samples from Kuala Lumpur, 9 water samples from Penang, 5 water samples from Melaka and 3 water samples from Kedah with different sources were collected. Out of 20 water samples from Selangor, 16 water samples were gram negative bacilli GNB which is demonstrated by gram staining. 4 water samples contained gram positive bacteria GPB. In 16 GNB, 6 showed positive in selective agars, which produced yellow greenish color fluorescent pigment in CA (Figure 1), beta hemolysis and greenish in color in BA (Figure 2), non-lactose fermented in MAC, which indicates Pseudomonas spp. (Figure 3). The remaining 10 water samples with GNB had no growth on CA, which indicates these GNB are not Pseudomonas species. In oxidase test, 8 out of 16 GNB showed positive result and the remaining 8 GNB showed negative results. The water samples with GNB were processed to biochemical test to identify the species of bacteria. Out of 16 GNB, 6 indicated P. aeruginosa, which showed negative results in Indole, Methyl red and Voges-prokauer. Positive results in Citrate and urease, motile in Motility test and Triple Sugar Iron test showed no gas and hydrogen sulphide H2S production, the remaining 10 indicated other than P. aeruginosa (Table 1).
In 15 water samples from Kuala Lumpur, 12 contained GNB 3 contained GPB. Among 12 water samples with GNB, 7 had Pseudomonas spp. grown in CA, BA, and MAC. The remaining 5 water samples with GNB had no growth on CA, which indicates these gram negative bacilli are not Pseudomonas spp. In oxidase test, 6 out of 12 GNB showed positive result, the remaining 6 GNB showed negative result In biochemical tests, out of 12 GNB, 7 indicated P. aeruginosa and the remaining 5 indicated other than P. aeruginosa.
Out of 9 water samples from Pinang, 6 had GNB, 3 had GPB. Out of 6 water samples with GNB, One had Pseudomonas spp. grown on CA, BA, and MAC. The remaining 5 water samples with GNB were not grown on CA, which indicates these GNB are not Pseudomonas spp. In oxidase test, 2 out of 6 GNB showed positive result, the remaining 4 GNB showed negative result. In biochemical tests, out of 6 GNB, 1 indicates P. aeruginosa whereas the remaining 5 indicates other than P. aeruginosa. Among 3 water samples from Kedah, 3 were GNB, but none were grown on CA, which indicates these GNB are not Pseudomonas spp. In oxidase test, 1 out of 3 GNB showed positive result, another 2 GNB showed negative result. In biochemical tests, all GNB indicates other than P. aeruginosa.
Lastly, among 5 water samples from Melaka, all of 5 contained GNB. In these 5 GNB, 1of Pseudomonas spp. was grown in CA (Figure 1) BA (Figure 2) and MAC (Figure 3). The remaining 4 GNB were not grown on CA, which indicates these GNB are not Pseudomonas spp. In oxidase test, 2 out of 5 GNB showed positive result, the remaining 3 GNB showed negative results. In biochemical tests, of 5 GNB, 1 was indicated P. aeruginosa, the other 4 showed other than P. aeruginosa.
Thus, among 52 water samples, 13 water samples were found positive for P. aeruginosa isolates. The rates of P. aeruginosa isolated from 13 water samples in different states of Malaysia showed 6 out of 20 water samples from Selangor were positive P. aeruginosa isolates, which was 46.1%. 5 out of 15 water samples from Kuala Lumpur were positive P. aeruginosa isolates which was 38.4 %, while 1 out of 9 water samples from Penang was positive P. aeruginosa isolates, which was 7.6%. 1 out of 5 water samples from Melaka was positive P. aeruginosa, which was 7.6% and lastly Kedah had not showed any positive P. aeruginosa isolates among 3 water samples.
Antibiotic susceptibility testing
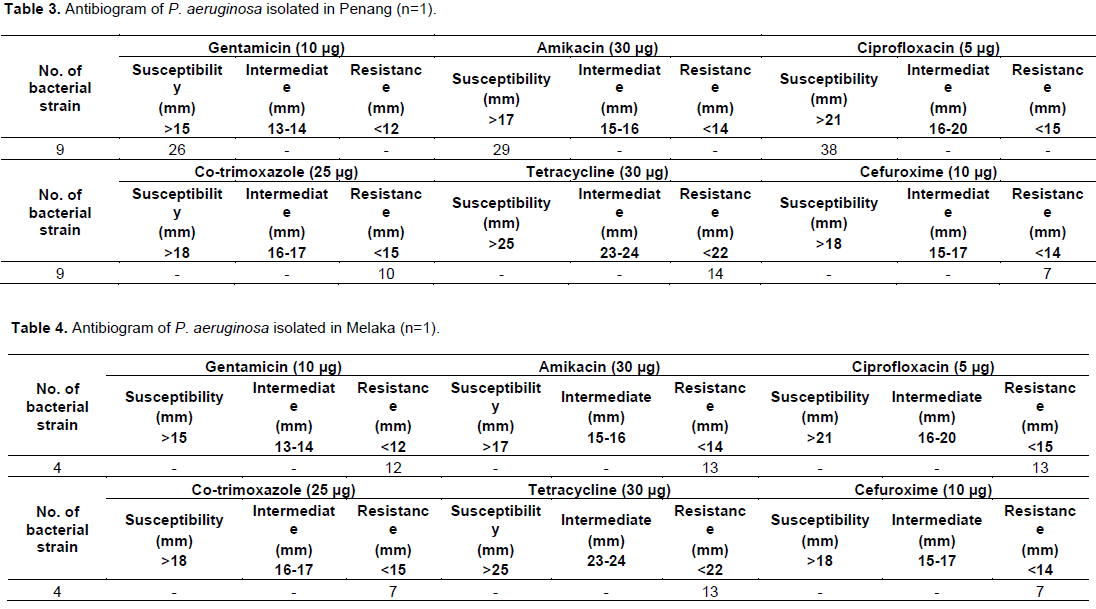
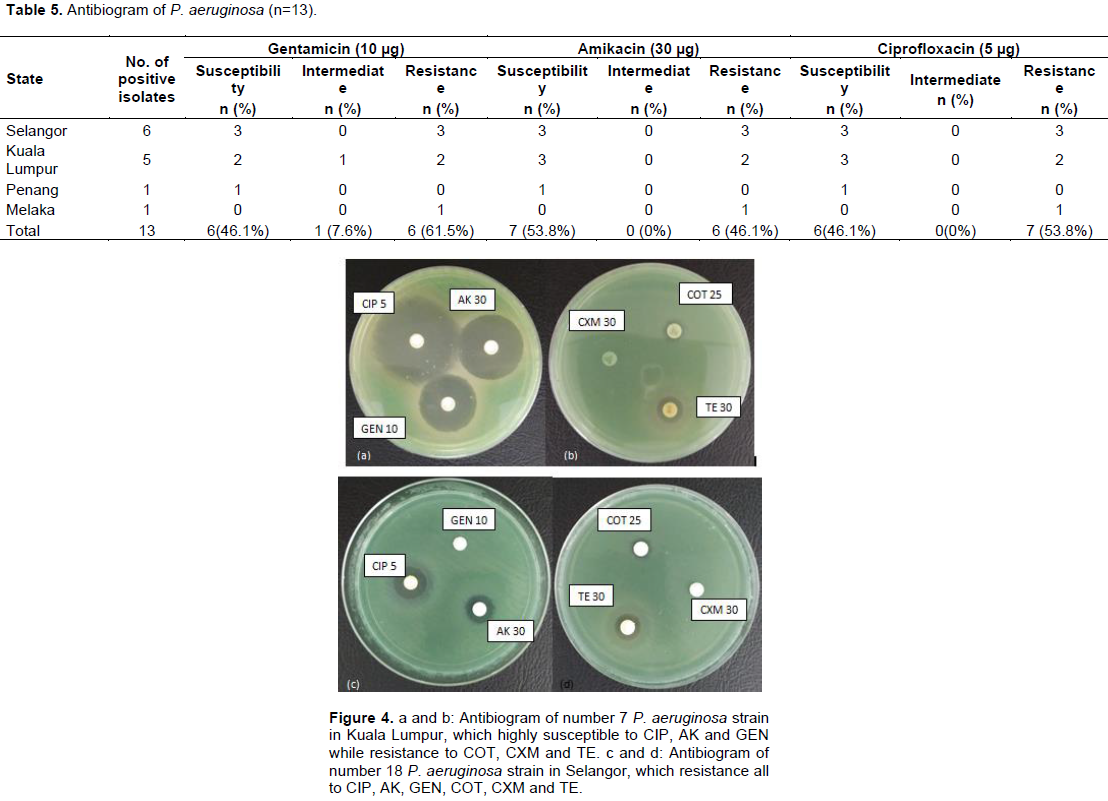
The antibiotic susceptibilities of the P. aeruginosa isolated in different states of Malaysia were as shown from Tables 2 to 5 and Figures 8 and 9. Out of 6 P. aeruginosa isolated from Selangor, number 7 P. aeruginosa isolate was highly susceptible to Gentamicin (GEN), Amikacin (AK) and Ciprofloxacin (CIP), Intermediate to Co-trimoxazole (COT), resistant to Tetracycline (TE) and Cefuroxime (CXM). Number 11 and 14 P. aeruginosa isolates were highly susceptible to Gentamicin, Amikacin and Ciprofloxacin, but resistance to Co-trimoxazole, Tetracycline and Cefuroxime. Whereas number 16, 18 and 19 P. aeruginosa isolates were resistant the entire antimicrobial disc test (Table 2b; Figure 4c and d). Of the 5 P. aeruginosa isolated from Kuala Lumpur number 4 and 7 P. aeruginosa isolates were highly susceptible to Gentamicin, Amikacin and Ciprofloxacin, resistant to Co-trimoxazole, Tetracycline and Cefuroxime (Figure 4a and b). Number 13 were intermediate to Gentamicin, susceptible to Amikacin and Ciprofloxacin. Number 5 and 14 P. aeruginosa isolates were resistant to all of the antimicrobial tests (Table 2a). The only one P. aeruginosa isolated from Penang was susceptible to Gentamicin, Amikacin and Ciprofloxacin, resistant to Co-trimoxazole, Tetracycline and Cefuroxime (Table 2b). P. aeruginosa isolated from Melaka was resistant to all of the antimicrobial drugs used. (Table 3) In addition, It has found that most of the P. aeruginosa isolates (n=13) were resistant to Co- trimoxazole, Tetracycline and Cefuroxime compared to Gentamicin, Amikacin and Ciprofloxacin (Table 4). The multidrug resistance P. aeruginosa isolates were indicated by resistant more than three antimicrobial drug resistant. Figures 7 and 8 showed antimicrobial susceptibility and resistance of P. aeruginosa isolates.
Hodge test
All 13 MDR P. aeruginosa isolates were tested for carbapenemase production. Among these 13 MDR P. aeruginosa 3 out of 6 from Selangor were positive of carbapenemase production, 2 out of 5 from Kuala Lumpur were positive of carbapenemase production and only one P. aeruginosa isolates from Penang and Melaka were positive. Thus, among 13 MDR P. aeruginosa isolates, 7 were carbapenemase positive and 6 were no carbapenease production (Table 5 and Figure 5).
Imipenem-EDTA combined disc test (CDT)
Out of the 13 MDR P. aeruginosa isolates, Selangor and Kuala Lumpur gave MBL positive, which were 5 out of 6, 4 out of 5 and only one from Melaka respectively. CDT had detected 10 MBL positive and only 3 MBL negative among the 13 MDR P. aeruginosa isolates. The MBL positive observed increase in EDTA with imipenem inhibition zone more than 7 mm in the imipenem alone (Table 6 and Figure 6).
Imipenem-EDTA double- disc synergy test (DDST)
Among 13 MDR P. aeruginosa isolates, 4 out of 6 from Selangor gave positive result, which showed the presence of synergistic zone of inhibition between two discs and indicated MBL production. 2 out of 5 from Kuala Lumpur showed MBL production. The only P. aeruginosa isolates from Melaka showed MBL production. However the only P. aeruginosa isolate from Penang showed negative result with no MBL production. Among 13 MDR P. aeruginosa isolates, 7 showed MBL production and 6 showed non-MBL production, which gave the same results after repeated tests (Table 7 and Figure 7).
Urban areas use clean water and produce large amounts of wastewater. Water is an important requirement in many industrial processes as well as hospital Corcoran et al. (2010). Antibiotics were used as a treatment for patient in hospitals. These antibiotics and their metabolites were excreted with urine and faeces and ended up in the wastewater stream (Chartier et al., 2014; Email et al., (2010). Hospital wastewaters were a source of bacteria with acquired resistance against antibiotics which has higher level compared with domestic wastewater Chartier et al. (2014). Industrial wastewater has the potential to be a highly toxic source of pollution if wastewater from industry drains directly into rivers and lakes without appropriate treatment (Corcoran et al., 2010). P. aeruginosa is a highly adaptable bacterium, they are able to grow well in water systems and have intrinsic antimicrobial resistance due to low outer membrane permeability as well as an extensive efflux pump system (Igbinosa et al., 2012). Therefore it increases the risk of health and environment.
Ministry of Health (2013) and Association of Private Hospitals of Malaysia (2016) showed that among the states in Malaysia, Selangor has the highest number of hospitals (18 hospitals including government and private hospital), followed by Kuala Lumpur (14 hospitals), while 12 hospitals in Penang, 9 hospitals in Kedah and 6 hospitals in Melaka. According to Makky et al. (2012), their study revealed that antibiotic resistance P. aeruginosa was isolated from hospital drains. Many studies on P. aeruginosa strains from clinical samples in hospitals were carried out (Butt et al., 2005; Aghamiri et al., 2014; Akya et al., 2015; Devi et al., 2015). In the present study, 52 water samples were collected from different states of Malaysia. 13 water samples were found positive for P. aeruginosa isolates. Among the P. aeruginosa isolates from various water sources in the different states of Malaysia, both Selangor and Kuala Lumpur (KL) have the highest rates of P. aeruginosa isolation compared to other states like Penang, Melaka and Kedah respectively. These 13 P. aeruginosa isolates were processed for antimicrobial susceptibility test. Out of these 13 P. aeruginosa isolation, 6 of P. aeruginosa isolation from Selangor indicated multi-drug resistance; 2 out of 5 of P. aeruginosa isolation from KL were MDR; P. aeruginosa isolates from Penang and Kedah did not show any MDR in antimicrobial susceptibility test; P. aeruginosa isolates from Melaka showed one MDR. Makky et al. (2012) mentioned that the development of resistant through mutation can also play an important role in development of β-lactam resistance.
In our study, Ciprofloxacin (5 µg), Gentamicin (10 µg), Amikacin (30 µg), Co- trimazole (25 µg), Tetracycline (30 µg), and Cefuroxime (30 µg) were used in antimicrobial susceptibility test. The 13 P. aeruginosa isolates were highly resistant to Tetracycline and Cefuroxime (100%) followed by Co-trimazole (92.3%), while moderate resistance with Ciprofloxacin 53.8%, Gentamicin and Amikacin (both 46.1%) respectively. According to Adesoji et al. (2015) P. aeruginosa isolates were more resistant to older generation antibiotics such as Sulfamethoxazole, Tetracycline, and Streptomycin than new generation antibiotics like Gentamicin, Kanamycin and Nalidixic acid. A recent study showed that P. aeruginosa isolates from clinical samples were found to be resistant to multiple-drug. The resistance level was 100% in Iran, 20.69% in Nepal, 19.6% in Malaysia (Nasreen et al., 2015). Akinpelu et al. (2014) stated that Pseudomonas strains from river were resistant to Gentamicin, followed by Co-trimazole, Tetracycline and Ciprofloxacin respectively. P. aeruginosa isolates from water sample in recent study (Nasreen et al., 2015) showed 93.7% isolates resistant to both Tetracycline and Gentamycin, 71.8% resistant to Co- trimoxazole and the P. aeruginosa isolates were found completely sensitive to Ciprofloxacin (100%). There are similarities between both studies, P. aeruginosa isolates are resistant to both Gentamicin and Co-trimoxazole but low resistant to Ciprofloxacin. In contrast, other study (Bhalerao et al., 2010) indicated P. aeruginosa isolates from clinical samples showed highly resistant to Amikacin (97.06%), followed by Ciprofloxacin (96.43%), Co-trimazole (95.24%), Gentamicin (85.7%) and Cefuroxime (75%). Based on these studies, it revealed P. aeruginosa isolates from water samples and clinical samples as well are highly resistant to Co-trimoxazole compared to Gentamicin and Tetracycline. Whereas, P. aeruginosa isolates from water samples are more susceptible to Ciprofloxacin than P. aeruginosa isolates from clinical samples, which showed high resistance (Bhalerao et al., 2010).
In this study, Hodge test detected 7 carbapenemase production, which gave 53.8% positive results, the number of positive results in Hodge test were same with DDST, which is 7 MBL production, 53.8%. While CDT detected 10 MBL positive 76.9% and only 3 MBL negative 23%. Based on the results among these phenotyping methods, CDT showed most sensitivity to Hodge test and DDST, yet Hodges test and DDST gave more specific results compared to CDT. Both Hodge test and DDST had similar results.
The metallo-β-lactamase producing P. aeruginosa were highly found in the Selangor followed by Kuala Lumpur. And the metallo-β-lactamase producing P. aeruginosa were detected from drain water followed by river. Selangor have more hospitals and industries than other states, therefore it may also increase the water contamination. Every water source should be tested for contamination with P. aeruginosa, especially metallo-β-lactamase-producing P. aeruginosa isolates, to prevent the infection caused by this organism. Water source should be properly filtered and treated to improve the water quality, and must be disinfected to prevent the emergence of these P. aeruginosa.
The authors have not declared any conflict of interests.
REFERENCES
|
Adesoji AT, Ogunjobi AA, Olatoye IO (2015). Molecular characterization of selected multidrug resistant Pseudomonas from water distribution systems in southwestern Nigeria. Ann. Clin. Microbiol. Antimicrob. 14(39):1.
Crossref
|
|
|
|
Aghamiri S, Amirmozafari N, Mehrabadi JF, Fouladtan B, Kafil HS (2014). Antibiotic Resistance Pattern and Evaluation of Metallo-Beta Lactamase Genes Including bla-IMP and bla-VIM Types in Pseudomonas aeruginosa Isolated from Patients in Tehran Hospitals. ISRN Microbiology. Volume 2014, Article ID 941507:6.
|
|
|
|
Akinpelu AT, Akinloye OM, Olukemi BC, Adegoke AE, Olayinka S (2014). Antibiotic resistant pattern of isolated bacteria from Obere River in Orile-Igbon, Oyo State, Nigeria. Afr. J. Microbiol. Res. 8(12):1318-1321.
Crossref
|
|
|
|
Akya A, Salimi A, Nomanpour B, Ahmadi K (2015). Prevalence and Clonal Dissemination of Metallo-Beta-Lactamase-Producing Pseudomonas aeruginosa in Kermanshah. Jundishapur J. Microbiol. 8(7):e20980.
Crossref
|
|
|
|
Arunagiri K, Sekar B, Sangeetha G, John J (2012). Detection and Characterization of Metallo-β-lactamases in Pseudomonas aeruginosa by Phenotypic and Molecular Methods from Clinical Samples in a Tertiary Care Hospital. West Indian Med. J. 61(8):778.
|
|
|
|
Association of Private Hospitals of Malaysia (2016). Hospitals/Medical Centers.
View
|
|
|
|
Bashir D, Thokar MA, Fomda BA, Bashir G, Zahoor D, Ahmad S, Toboli AS (2011). Detection of metallo-beta-lactamase (MBL) producing Pseudomonas aeruginosa at a tertiary care hospital in Kashmir. Afr. J. Microbiol. Res. 5(2):164-172.
|
|
|
|
Bhalerao DS, Roushani S, Kinikar AG, Akhter (2010). Study of Metallo-beta lactamase producing Pseudomonas aeruginosa in Pravara Rural Hospital. Pravara Med Rev 5(3):16-19.
|
|
|
|
Biradar S, Roopa C (2015). Prevalence of Metallo-Beta-Lactamase Producing Pseudomonas aeruginosa and its antibiogram in a tertiary care centre. Int. J. Curr. Microbiol. Appl. Sci 4(9):952-956.
|
|
|
|
Butt T, Usman M, Ahmad RM, Saif I (2005). Emergence of Metallo-beta- Lactamase producing Pseudomonas aeruginosa in Pakistan. J. Pak. Med. Assoc. 55(7):302.
|
|
|
|
Chartier Y, Emmanuel J, Pieper U, Prüss A, Rushbrook P, Stringer R, Townend W, Wilburn S, Zghondi R (2014). Safe management of wastes from health-care activities. 2nd ed. Switzerland: World Health Organization.
|
|
|
|
Chaudhari MS, Javadekar TB, Ninama G, Pandya N, Damor J (2011). A study of metallo-beta-lactamase producing pseudomonas aeruginosa in clinical samples of S.S.G hospital. Natl. J. Med. Res. 1(2):60-63.
|
|
|
|
Chen CT, Tsay JD, Yen MC, Matsumoto J (2013). The Winter Rainfall of Malaysia. J. Clim. 26(3):936-58.
Crossref
|
|
|
|
Corcoran E, Nelleman C, Bake E, Bos R, Osborn D,Savelli H (2010). Sick Water? The central role of wastewater management in sustainable development. UNEP/Earthprint.
|
|
|
|
Devi PV, Reddy PS, John MS (2015). Prevalence of Metallo-Lactamases Producing Pseudomonas aeruginosa among the Clinical isolates: A study from tertiary care hospital. Int. J. Curr. Microbiol. Appl. Sci. 4(4):955-996.
|
|
|
|
Email VD, Tamhankar AJ, Khandal RK, Sen S, Aggarwal M, Marothi Y, Iyer RV, Tonderski KS, Lundborg CS (2010). Antibiotics and antibiotic- resistant bacteria in waters associated with a hospital in Ujjain, India. BMC Public Health 10(1):414.
Crossref
|
|
|
|
Georgios M, Egki T, Effrosyni S (2014). Phenotypic and Molecular Methods for the Detection of Antibiotic Resistance Mechanisms in Gram Negative Nosocomial Pathogens. Trends Infect. Dis. InTech,
Crossref
|
|
|
|
Hallin M, Deplano A, Roisin S, Boyart V, Ryck RD, Nonhoff C, Byl B, Glupczynski. Y, Denisa O (2012). Pseudo-Outbreak of Extremely Drug-Resistant Pseudomonas aeruginosa Urinary Tract Infections Due to Contamination of an Automated Urine Analyzer. J. Clin. Microbiol. 50(3):580-582.
Crossref
|
|
|
|
Hussain M, Prasad Rao TVD (2013). Effect of Industrial Effluents on Surface Water Quality - A Case Study of Patancheru, Andhra Pradesh, India. Curr. World Environ. 8(3):445-454.
Crossref
|
|
|
|
Igbinosa EO, Odjadjare EE, Igbinosa IH, Orhue PO, Omoigberale MNO, Amhanre NI (2012). Antibiotic Synergy Interaction against Multidrug-Resistant Pseudomonas aeruginosa Isolated from an Abattoir Effluent Environment. Sci. World J. 2012:308034.
Crossref
|
|
|
|
James Cappuccino, Natalie Sherman (2007).Microbiology: A Laboratory Manual, Benjamin Cummings; 8 edt.
|
|
|
|
Kali A, Srirangaraj S, Kumar S, Divya HA, Kalyani A, Umadevi S (2013). Detection of metallo-beta-lactamase producing Pseudomonas aeruginosa in intensive care units. Aust. Med. J. 6(12):686-693.
Crossref
|
|
|
|
Kumar SH, De AS, Baveja SM, Gore MA (2012). Prevalence and Risk Factors of Metallo β-lactamase Producing Pseudomonas aeruginosa and Acinetobacter species in Burns and Surgical Wards in a Tertiary Care Hospital. J. Lab. Physicians 4(1):39-42.
Crossref
|
|
|
|
Lambert PA (2002). Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J. R. Soc. Med. 95:22-26.
|
|
|
|
Makky EA, Ibrahim MM, El-Gamal MS (2012). Survey of Hospital Drains Antibiotic Resistant Bacteria for Hygiene and Health Care. International Conference on Chemical, Biological and Medical Sciences (ICCBMS'2012) August 25-26, 2012 Kuala Lumpur (Malaysia), Pp. 1-4.
|
|
|
|
Manoharan A, Chatterjee S, Mathai D (2010). Detection and characterization of metallo beta lactamases producing Pseudomonas aeruginosa. Ind. J. Med. Microbiol. 28:241-244.
Crossref
|
|
|
|
Ministry Of Health Malaysia (2013). List of Government Hospitals.
View
|
|
|
|
Nasreen M, Sarker A, Malek MA, Ansaruzzaman Md, Rahman M (2015). Prevalence and Resistance Pattern of Pseudomonas aeruginosa Isolated from Surface Water. Adv. Microbiol. 5(1):74.
Crossref
|
|
|
|
Pellegrini C, Mercuri. PS, Celenza G, Galleni M, Segatore B, Sacchetti E, Volpe R, Amicosante G, Perilli M (2009). Identification of blaIMP-22 in Pseudomonas spp. in urban wastewater and nosocomial environments: biochemical characterization of a new IMP metallo-enzyme variant and its genetic location. J. Antimicrob. Chemother. 63(5):901-908.
Crossref
|
|
|
|
Qu TT, Zhang JL, Wang J, Tao J, Yu YS, Chen YG, Zhou JY, Li LJ (2009). Evaluation of Phenotypic Tests for Detection of Metallo-β-Lactamase- Producing Pseudomonas aeruginosa Strains in China. J. Clin. Micro. 47(4): 1136- 1142.
Crossref
|
|
|
|
Quick J, Cumley N, Wearn CM, Niebel M, Constantinidou C, Thomas CM, Pallen MJ, Moiemen NS, Bamford A, Oppenheim B, Loman NJ (2014). Seeking the source of Pseudomonas aeruginosa infections in a recently opened hospital: an observational study using whole-genome sequencing. BMJ Open 4(11):e6278.
Crossref
|
|
|
|
Slekovec C, Plantin P, Cholley P, Thouverez M, Talon D, Bertrand X, Hocquet D (2012). Tracking down Antibiotic-Resistant Pseudomonas aeruginosa Isolates in a Wastewater Network. PLoS One 7(12):e49300.
Crossref
|
|
|
|
Upadhyay S, Joshi SR (2015). TEM mediated extended spectrum cephalosporin resistance in clinical & environmental isolates of Gram negative bacilli: A report from northeast India. Ind. J. Med. Res. 142:614-617.
Crossref
|