ABSTRACT
Bacterial plant pathogens belonging to the Xanthomonas genus are adapted to their host plants and are not known to colonize other environments. Xanthomonas colonize host parts such as leaves, stems and roots before entering vascular tissues and engaging in an invasive pathogenic phase. These bacterial strains have evolved strategies to adapt to life in this environment. The host-pathogen interactions of Xanthomonas vasicola (Xv) need to be well understood to properly map the target genes in the host and pathogen so as to understand the mechanism of resistance. Genotypic characterization, based on the analysis of restriction fragment length polymorphism of virulence factor fragment products was performed on members of the X. vasicola pv. musacearum (Xcm) and X. vasicola pv. vasculorum (Xvv) from varying geographical locations. The study showed that Xcm and Xvv are different from each other based on amplification of virulence factors within fragments of their DNA. Bacterial strains of similar species can have unique Type four pili (Tfp) and Tfp pilus assembly protein PilF a fimbrial biogenesis protein was amplified in all Xanthomonas strains except NCPPB1131 only. Type III effector protein RipT was confirmed to be present in all strains of Xcm and Xvv but not NCPPB1131 and NCPPB1132. All the Xcm and Xvv strains under test yielded bands of type III effector HopAF1 except Xvv206, NCPPB1131 and NCPPB1132. YopJ type III secretion system effector protein hybridizes in DNA of all Xcm strains tested but not in NCPPB1131 or NCPPB1132. This study confirmed the predicted presence or absence of virulence factors especially effectors across bacterial strains and within strains of the same species and other clusters conserved in gram negative bacteria.
Key words: Banana, effectors, pathogen-host, Xanthomonas wilt, Xanthomonas campestris, Xanthomonas vasicola.
Banana production in Eastern Africa is on the decline due to diseases particularly Banana Xanthomonas Wilt (BXW) (Kubiriba et al., 2012; Tushemereirwe et al., 2004). This correlates to 80% yield loss and thus 32% loss in household income. Currently the most effective control methods are cultural based and the deployment of resistance would be most ideal and less expensive on time and resources. Efforts are being made to develop transgenic BXW resistant banana varieties using Hypersensitive Response Assisting Protein (Hrap) or Plant Ferredoxin like Protein (Pflp) gene originated from sweet pepper (Capsicum annuum). The interaction of Xcm and its banana host needs to be well understood to properly map the target genes in host and also understand the mechanism of resistance to support future control programs (Tripathi et al., 2010).
Xanthomonas is a large genus of Gram-negative bacteria that cause disease in hundreds of plant hosts, including many economically important crops. For bacteria to adapt specifically to their hosts, they sense favorable environmental stimuli and then they move toward favorable conditions. Bacteria have evolved receptors and sensors in their cell walls to detect chemical and environmental signals such as the presence of chemo-attractants and chemo-repellents (Mhedbi-Hajri et al., 2011). Pathogenic species and pathovars within species show a high degree of host plant specificity and exhibit tissue specificity, invading either the vascular system or the mesophyll tissue of the host. Plant-pathogenic Xanthomonas pathovars require a type three secretion system (TTSS) to secrete and translocate effector proteins in order to cause disease. The repertoire of effectors can vary between species and strains within species and is believed to be a key determinant in the host range of a given pathogen (Baltrus et al., 2011). The draft genome analysis of the two bacteria species (Xcm4381 and Xvv702) revealed several genetic differences between the two strains that might be important for host specificity, virulence and epiphytic fitness, including differences in the repertoires of secreted and translocated effector proteins, Tfp and enzymes for lipopolysaccharide biosynthesis (Studholme et al., 2010). They both encode homologues of the candidate T3SS effectors including AvrBs2, AvrGf1, HopW1; HopAF1; PilvD; RipT; YopT-like cysteine protease and a variety of homologues of XopF, XopK, XopL, XopN, XopP, XopQ, XopR, XopX, XopZ, XopA, XopB, XopG, XopH, XopI, XopY, XopAA, XopAD, XopAE and XopAK, which are conserved in a subset of Xanthomonas genomes. However Xcm4381 encodes two predicted YopJ-like C55 cysteine proteases that are absent from Xvv702, whereas Xvv702 encodes a protein XopAF (also known as AvrXv3) which is absent from Xcm4381, but shares 35% identity with the HopAF1-like genes. Such differences in effector repertoires are said to be significant for host adaptation.
Lipopolysaccharide (LPS), which are produced by Gram-negative bacteria, are powerful activator of inherent immune responses. The lipopolysaccharide locus in Xvv702 showed no significant sequence similarity to that of Xcm4381. The respective Type IV pilus (Tfp) cluster showed little sequence similarity between proteins, respectively encoded on the Xvv702 and the Xcm4381 TFP clusters. An 8-kb gene cluster in Xcm4381 is reported to encode TFP components FimT, PilV, PilW, PilX, PilY1 and PilE. A different gene cluster in Xvv 702 encodes homologues of TFP components FimT, PilE, PilY1, PilW and PilV. These differences might be adaptive for attachment to and motility on different plant surfaces.
There is need to clearly identify and understand the infection process applied by Xcm and Xvv so as to propose pathogen protein targets for suppressing the motility, mode of host plant surface identification, vascular rapid multiplication and evasion of host plant resistance mechanism. This study therefore aimed at experimentally validating presence or absence of predicted virulence factors in different strains of Xcm, Xsp.and Xvv.
Bacterial culture conditions
Bacterial cultures of strains in Table 1 were obtained from National Collection of Plant Pathogenic Bacteria (NCPPB) at the Food and Environmental Research Agency (FERA, York, UK). The samples were received on filter paper in glass vials, recovered through streaking on solid Kings Broth (KB). The KB was prepared using 10 g Peptone meat, 10 g N-Z casein; 1.5 g MgSO4-7H2O; 1.5g K2HPO4; 12.6 g glycerol to a liter of dH2O and stabilized at pH 7.2. Cultures were further purified by re-streaking onto new KB in a fixed incubator for 12 h. A single loop colony from each strain was transferred into 10 ml liquid KB and grown at 28°C in a shaking incubator at 150 rpm overnight. All of the isolates used in this study were then put in glycerol stock and kept at -20°C for further use during the experiment and -80°C for long term storage.
Total genomic DNA extraction from cultures
Genomic DNA from each of the strains was extracted using modified protocols by Mahuku (2004). Overnight cultures were centrifuged at 4000 rpm 4°C for 10 min to obtain a cell pellet, and supernatant was poured off and the pellet dried. Cells were suspended and dissolved in 2 ml of TE buffer (25 mM Tris Hcl pH 8.0, 10 mM EDTA). 300 µl cells were lysed by adding 12 µl lysozyme 20 mg/µl. 1.5 µl RNase 10 mg/µl were then added and mixed by inverting 10 times and incubated at 25°C for 10 min. 17 µl 10% SDS were then added and mixed by inverting 10 times then incubated on ice for 5 min. Proteins were pelleted by adding 170 µl 8 M ammonium acetate (not pH), mixed by inverting 10 times, vortexed 20 s and spanned 4°C at 4000 rpm for 30 min. The supernatant was carefully pippeted off into clean tubes avoiding the pellet and membrane formed on the surface. DNA was pelleted by adding 0.75% Vol Isopropanol, mixed by inverting until a ball of DNA was visible; then centrifuged at 4°C, maximum speed for 10 min. The supernatant was pippeted off and air dried for 5 min. The pellet was washed with 100 µl 70% ethanol; centrifuged for 1 min maximum speed, the ethanol pipetted off, spinned short burst and pipette off excess ethanol; air dry pellet 5 mins. The pellet was then dissolved in 200 µl TE, incubated in 65°C water bath for 30 min to 1 h. The quality and quantity of DNA was then quantified using a NanoDrop® ND-1000 (NanoDrop Technologies USA,
www.nanodrop.com). Agarose gel electrophoresis of each sample DNA was run on 0.8% agarose gel for 45 min at 100 V.
Primer design and validation of putative virulence factors
The primers for 26 virulence factors were designed using Primer3 (Koressaar and Remm, 2007; Untergasser et al., 2012) to amplify the full length gene. The Primer-Search program was used to confirm the specificity of the primers (cut off of 20% mismatch) against the sequenced Xcm4381 and Xvv702. Each 25 µl of PCR mixture contained 1.5 mM MgCl2, 0.2 µM forward and reverse primers, PCR buffer (Invitrogen, Carlsbad, CA), 0.2 mM each deoxynucleoside triphosphate (dNTP), 0.5 units Taq polymerase (Invitrogen, Carlsbad, CA), and 10 ng DNA. After an initial denaturing step, PCR was conducted for 28 cycles of 30 s at 94°C, 30 s at 55°C, and 70 s at 72°C. The primers were tested on DNA of Xcm4381 and Xvv702 and those validated used to amplify bacterial strains in Table 1.
Cluster analysis
The data was converted as follows: Absence of a band (-) changed to (0) and presence of a band (+) changed to (1) before performing a hierarchical cluster analysis. The distance was measured using the Binary Squared Euclidean distance and separated by the between groups method. A dendogram was then produced by the SPSS 16 Edition program.
Primers used in the study are detailed in Table 2 and amplified with DNA from test bacterial strains whose response is listed in Tables 3 and 4. Virulence factors were categorized into lipopolysaccharides (LPS), type iv pilus proteins (Tfp) and type three secretion system (TTSS) proteins for analysis across 8 strains of Xcm, 2 strains of NCPPB and 3 Xvv strains. Only primers that amplified with DNA of the selected strains are given against a 1000 bp ladder. The amplification of DNA was scored as present or absent for the different strains in the study, converted to binry data (0 for -) and (1 for +). The converted data was subjected to cluster analysis using a two-step approach in SPSS 16.0 (SPSS, 2008). The binary data was then clustered using the square Euclidean distance method.
Lipopolysaccharides variation in selected Xcm and Xvv strains
Xcm strains had lipopolysaccharides
ABC transporter permease/ATP-binding protein (permease); S-adenosyl-L-methionine (SAM) dependent methyltransferase, truncated O-antigen biosynthesis protein and hypothetical protein ZP_06489485 amplified but had no product with NCPPB1131, NCPPB1132, Xvv206, Xvv1326 and Xvv1381 strains (Figure 1).
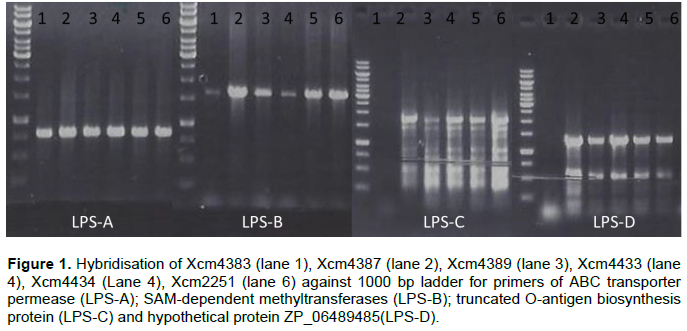
Predicted membrane protein (YP_007652590) was present in all Xcm strains (LPS-E) and also in NCPPB1132, Xvv 1326 and 1381 but not observed in Xsp 1131 and Xvv206 (LPS-). Nicotinamide adenine dinucleotide (NAD) dependent epimerase was observed in Xcm4387, Xcm4389, Xcm4433, Xcm 4434, Xcm2251, Xcm2005, Xcm4392, NCPPB1131, NCPPB1132 and unexpectedly in Xvv206, Xvv1326 and Xvv1381, interestingly we did not observe it in Xcm 4383 (LPS-F). The protein, indolepyruvate ferredoxin oxidoreductase (LPS-G) was amplified in all the Xcm strains except Xcm4392 and although expected in Xvv strains, it was not amplified (Figure 2).Xcm4383, Xcm2005, Xcm4392, NCPPB1131, NCPPB1132 and all the Xvv strains did not amplify the lipopolysaccharide biosynthesis protein (Figure 3). Ferredoxin oxidoreductase (Xvv702 based) hybridised only in Xcm4387, Xcm4433, Xcm4434 and Xcm2251 strains (LPS-K); also in Xcm2005, Xvv206 and Xvv1326. Another protein short chain dehydrogenase/reductase was amplified in Xcm4387, Xcm2005 and Xcm4392 just like was expected in Xvv206, Xvv1326 and Xvv1381; the other Xcm strains including NCPPB1131 and NCPPB1132 did not yield the protein (Figure 3).
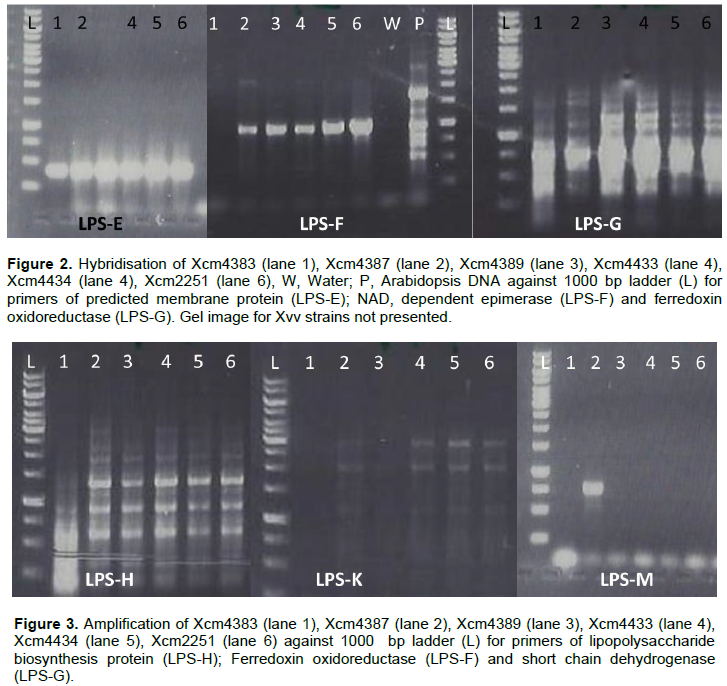
Strain Xcm2005 unexpectedly amplified putative transmembrane GtrA and GDP-mannose 4,6-dehydratase protein just like Xvv206, Xvv1326 and Xvv1381, the rest of the test strains did not yield any strands of the protein (Figure 4).
Type IV pili of Xcm, Xvv and Xsp strains
Here we show that bacterial strains of similar species can have unique Tfp. Primers specific to Xcm4381, amplified a unique product for Xcm2251 of size 1500 kb, whereas Xcm4383, Xcm4387, Xcm4389, Xcm4433, Xcm4434, amplified similar product except NCPPB1131, NCPPB1132 (Tfp-A). Tfp pilus assembly protein PilE specific to Xvv702 was amplified in Xcm2005, Xvv1326 and Xvv1381 DNA but tested negative in DNA of the other Xanthomonas test strains (Tfp-B). Tfp pilus assembly protein FimT was amplified in DNA of Xvv1326 and Xvv1381 but not Xvv206
nor any of the other strains (Tfp-C) (Figure 5).
Tfp pilus assembly protein PilF a fimbrial biogenesis protein was amplified in all Xanthomonas strains except NCPPB1131 only (Figure 6).
Tfp pilus assembly protein, PilV(Tfp-F) and PilW (Tfp-G) were not amplified in NCPPB1131/1132, Xvv1326 and Xvv1381 but amplified for the rest of the Xanthomonas strains under test (Figure 7).
DNA of Xcm4383, NCPPB1131 and NCPPB1132 did not amplify with Tfp pilus assembly protein PilX primers although the other strains had products as expected (Figure 8).
PilY1 a Tfp pilus assembly protein (tip-associated adhesion) was amplified in DNA of Xcm4387, Xcm4433, Xcm2251, Xcm2005, Xcm4392, and Xvv206, whereas no product was observed for strains Xcm4383, Xcm4389, Xcm4434, NCPPB1131, NCPPB1132, Xvv1326, and Xvv1381 (Figure 9).
Type three secretion system (TTSS) effectors
In this study we confirm that indeed there can be differences in the effectors across bacterial strains and within strains of the same species and other clusters conserved in gram negative bacteria. Type III effector protein RipT was confirmed to be present in all strains of Xcm and Xvv but not NCPPB1131 and NCPPB1132 (RipT). All the Xcm and Xvv strains under test yielded bands of type III effector HopAF1 except Xvv206, NCPPB1131 and NCPPB1132 (Figure 10).
Another type III effector HopW1 was amplified in all Xcm and Xvv strains tested except Xcm4383, NCPPB1131 and NCPPB1132 (Figure 11).
YopJ type III secretion system effector protein amplifies in DNA of all Xcm strains tested but not in NCPPB1131, NCPPB1132 and the Xvv strains tested (Figure 12). Xvv702, Xvv1326 and Xvv1381 strains yielded the only products for type III effector protein XopAF, with no amplification in any of the other Xanthomonas strains under test (results not presented).
Clustering of virulence factors
Cluster analysis of the presence or absence of virulence factors in bacterial strains separated the factors into two major clusters (Figure 13). The putative transmembrane GtrA, GDP-mannose 4,6 dehydratase, Tfp Pilus protein PilE, Tfp pillus assembly protein FimT, TTSS effector protein XopA and short chain dehydrogenase are in a unique cluster from the others assessed. These were mainly amplified in the Xvv strain and not in Xcm strains except for Xcm2005.
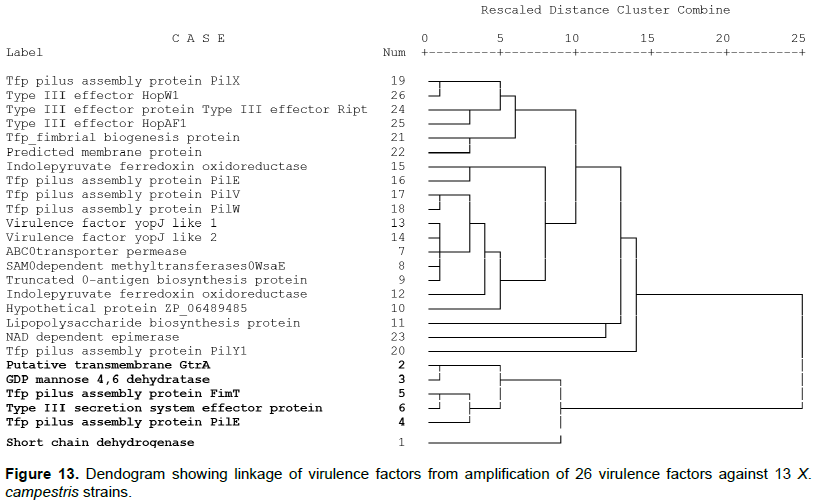
The cluster analysis further indicated that Xcm, Xvv strains and NCPPB did not cluster together (Figure 14). This is similar to observations made by Wasukira et al. (2012). Among the Xcm cluster strains Xcm4383 from Uganda clusters oni ts own withint he major group, another unique clustering is Xcm2005 and Xcm4392 that are further from the other Xcm strains. However the NCPPB1131 and NCPPB1132 were generally grouped with the Xvv strains. NCPPB1131 and NCPPB1132 form a separate sub cluster while Xvv1326 and Xvv1381 sub cluster together and in this major cluster Xvv206 from maize is further apart from the others.
Type four pili in Xcm and Xvv
In this study we have shown that Xcm and Xvv strains under test differ in their Tfp composition as was earlier predicted by Studholme et al. (2010). We experimentally confirmed that the structural subunit PilE is present in all the strains of Xanthomonas under test however it was not detected in NCPPB1131 and NCPPB1132. These strains do not possess the TTSS and are not related to Xcm nor Xvv (Studholme et al., 2011). PilE were able to amplify a product in Xcm2005 (Enset), Xvv1326, Xvv1381 (sugar cane) and no product was observed for Xvv206(maize) thus indicating a possible loss in the gene locus. The ability of pathogenic bacteria to infect plants involves more than their ability to form the Hrp-TTSS and transfer virulence effectors. Qian et al. (2005) reported that infection is initiated with bacterial attachments to and colonization of host tissues via surface structures and appendages. Attachment of plant pathogenic bacteria to host specific surfaces/cells is necessary for colonization of host tissue and is mediated by surface-exposed adhesins, which generally behave as lectins, recognizing oligosaccharide residues of glycoprotein or glycolipid receptors on the host cell (Pizarro-Cerda and Cossart, 2006; Kline et al., 2009). Pili are implicated in crucial host-pathogen interactions, colonization, tropism determination, biofilm formation, and invasion and signaling events. Tfp is said to contribute to the optimal establishment, colonization, and spread of vascular bacteria pathogens via the plant xylem vessels. In Xanthomonas, type IV pili may be associated with the establishment of an aggregated bacterial population necessary to counteract the turbulent environment of the xylem, facilitating its adherence to the vessels in conjunction with other components, such as exopolysaccharides (Sluys et al., 2002). Tens of genes are involved in TFP synthesis and regulation, with the majority of them being generally named pil/fim genes. Moreira et al. (2004) reports of the presence and divergence of copies of pilE-fimT cluster in ancestors of xanthomonadaceae is consistent with the findings of this study where fimT was amplified in only the Xvv1326 and Xvv1381 strain from sugarcane. PilV is reported to be essential for pilus mediated cell adherence. The involvement of minor pilins (that is, PilE, PilV, PilW, PilX, FimT) in pilus assembly may play roles in priming of pilus extension or prevention of pilus retraction; in control of pilus length; or in pilus-specific functions including adherence, transformation competence or motility (Dunger et al., 2014; Giltner et al., 2012). The putative type IV pilus protein PilY1 is likely important for attachment to surfaces. Moreira et al. (2004) reported a cluster pilE-pilY1-pilX-pilW-pilV-fimT which was common to both Xylella fastidiosa and X. axonopodis pv. citri. In this study it was experimentally confirmed that strains of the Xcm and Xvv may vary in clusters of Tfp. The roles of various Tfp between Xcm and Xvv should be further investigated for their interaction with the host and non-host(s).
Presence lipopolysaccharides in Xcm and Xvv strains
LPS are referred to as endotoxins and are known to play a wide range of roles during bacterial infection (Munford and Varley, 2006; Todar, 2014; Volk, 1966). LPSs share a common structure for all Gram-negative bacteria composed of a membrane-anchored phosphorylated and acylated 1–6-linked glucosamine (GlcN) disaccharide, named lipid A, to which a carbohydrate moiety of varying size is attached (Casabuono et al., 2011). The latter may be divided into a lipid A, proximal core oligosaccharide and a distal O-antigen, whose presence or absence determines the smooth or rough appearance of the bacterial colony. Studholme et al. (2010) had predicted that lipopolysaccharide locus in Xvv702 and Xcm4381 were not significantly similar which has been experimentally confirmed by this study. According to Wasukira et al. (2014), further genome sequencing did not reveal significant variation in this locus among isolates of Xvv. However, a predicted membrane protein was detected in strains of Xvv1326 and Xvv1381 but not Xvv206 which was isolated from maize unlike the former isolated from sugar cane. This was confirmed by Wasukira et al. (2014) who reported that the LPS structure of Xvv702 and Xvv206 were similar. LPSs apparently play diverse roles in bacterial pathogenesis of plants. As major components of the outer membrane, they are involved in the protection of bacterial cells, contributing to reduce the membrane permeability and thus allowing growth of bacteria in the unfavorable conditions of the plant environment. LPSs can be recognized by plants to elicit or potentiate plant defense-related responses (Casabuono et al., 2011; Desaki et al., 2006; Nam, 2001; White et al., 1996). It has also been inferred by Ssekiwoko et al. (2006) and Tripathi and Tripathi (2009) that X. vasicola pv. musacearum spread is limited to the xylem vessels and the wilting symptoms are as a consequence of large amounts of exopolysaccharides blocking water flow in the plants. One of the most widely studied effects of LPSs on plant cells is their ability to prevent the hypersensitive response (HR) induced in plants by avirulent bacteria (Mohammed, 2015; Newman et al., 2000). Newman et al. (2000) were able to show that LPS pre-treatment prevents the hypersensitive reaction caused by strains of Xanthomonas campestris pv. vesicatoria carrying the avirulence gene avrBs1 (a gene-for-gene interaction) and by X. campestris pv. campestris (a non-host interaction). X. axonopodis pv. citri 306 shares 93% nucleotide sequence similarity with Xcm4381 (Studholme et al., 2010) and Casabuono et al. (2011) suggested that the O-antigen region of X. axonopodis pv. citri 306 LPS could be involved in the innate immunity of citrus. Horizontal gene transfer events (HGT) are frequently observed in genomic regions that encode functions involved in biosynthesis of the outer membrane located lipopolysaccharide (LPS). Also different strains of the same pathogen can have substantially different LPS biosynthetic gene clusters as has been shown for NAD dependent epimerase and lipopolysaccharide biosynthesis protein in this study. This can be attributed to the advantage in evading the host immune system since LPS is highly antigenic. Although LPS has been suggested as a potentiator of plant defense responses, interstrain variation at LPS biosynthetic gene clusters has not been reported for any plant pathogenic bacterium. Aritua et al. (2008), study found that although banana and sugarcane strains were of similar phylogenic group, their host ranges were different, since only the X. campestris pv. musacearum pathovars could cause disease in banana (Studholme et al., 2010; Wasukira et al., 2012). This is consistent with the observation that lipopolysaccharide (LPS) O antigen is pathovar/strain-specific and may be involved in host-range selection and pathogenicity by acting as a barrier against plant toxins (da Silva et al., 2002).
Presence of Type III secretion system effectors (TTSS)
TTSS is defined as a multi-subunit protein apparatus that is used to secrete or inject effector proteins which contribute to interactions with eukaryotic cells (Costa et al., 2015; Peeters et al., 2006). Xanthomonas species genomes possess 2 pathogenesis associated gene clusters that include hypersensitive response and pathogenicity (hrp) and the gum genes that encode synthases for extracellular polysaccharides xanthan. These pathogens show high level of host plant specificity inclusive of tissue specificity. They invade either the xylem elements of the vascular system or the intercellular spaces of the mesophyll. Differences in the complement of effectors between different Xanthomonas strains of the same pathovar may determine host specificity at the cultivar level. Effector recognition may also underpin the host range restriction of pathovars within a species and of the species themselves to particular plants (Buttner and He, 2009; Coburn et al., 2007). In this study we were able to confirm that the genes RipT, HopAF1 and YopJ-like1 and YopJ-like2 were amplified within all Xcm strains under test. It was also observed that Xvv206, Xvv1326 and Xvv1381 did not yield any product for YopJ-like proteins. Homologs of the Yersinia virulence effector YopJ are found in both plant and animal bacterial pathogens. These YopJ family members act as cysteine proteases which is required for inhibition of the mitogen-activated protein kinase (MAPK) and nuclear factor kB (NF-kB) signaling for induction of localized cell death in plants (Orth et al., 2006). Although effector protein hopW1 and XopAF (AvrXv3) was amplified in Xvv1326 and Xvv1381, it was not detected in Xvv206. Jalan et al. (2013) in characterizing pathotypes of X. citri subsp. Citri showed that XopAF was present in Xcaw12879 but not XccA306 thus contributing to virulence in Xcaw. These observations are variable with predictions of Studholme et al. (2011) where NCPPB1131 was shown to be closely related to X. saccharum and also X. albilineans. However the strains lacked the Hrp TTSS of Xcm, and yet NCPPB1131 and NCPPB1132 possess a gum gene cluster similar to Xcm. This confirms the prediction by Studholme et al. (2010) who had indicated that genome of Xvv702 (sugar cane) encodes different protein compared to Xcm4381 (musa). Kvitko et al. (2009) proposed that unraveling functional redundancy among effectors can ease the study of individual effectors and elucidation of functional overlaps should help us understand how the various effectors in a repertoire may function as a system in hosts.
These results provide direction for functional studies on host specificity of plant pathogenic bacteria by using variable strains to study the role of single or combination of virulence factors/genes in the interaction with plants. Ali et al. (2013) recommend that comparative genomic analysis of intra-species by multiple strains is a modern trend for studying bacterial pathogens. These results will provide targets for insights into the molecular nature of virulence and host specificity; advance understanding of the dynamics responsible for banana Xanthomonas wilt epidemic development.
In conclusion the presence and or absence of virulence determinants is required to understand the mechanisms used by bacterial pathogens to establish infection. The construction of gene disruption mutants and their individual in vivo phenotype analysis is a common approach for the functional characterization of targeted genes. We used PCR-based screening to detect presence or absence of predicted virulence factors which is a fast and simple technique. This can be a first step towards characterization of predefined target genes from strains of X. campestris pv. musacearum and X. vasicola pv. vasculorum. This study has indicated as predicted that some of the predicted virulence factors are conserved within species whereas others can vary within species. The two specie strains also showed difference in the Tfp whose function is to aid in motility and attenuation to the host and thus tissue specificity. A management strategy for the disease can be based on genes which target the Tfp cluster. Lipopolysaccharides which act as PAMP are conserved within the Xcm and also vary with that found in Xvv a factor that explains the effect on host and non-host of each bacterium. Among the TTSS that were screened, XopAF was never amplified in Xcm strains, RipT, HopAF1 and YopJ-like1/2 were amplified in all Xcm. The correct identification of novel secreted effectors using protein sequence is a first step towards a more complete characterization of the complex pathogen-host interaction. This study has provided an indication of target virulence factors that can be used in further understanding the Xcm/Xvv interaction with the banana host especially targets for functional analysis of the virulence factors.
The authors have not declared any conflict of interests.
REFERENCES
|
Ali A, Soares SC, Barbosa E, Santos AR, Barh D, Bakhtiar SM, David W (2013). Bacteriology & Parasitology Microbial Comparative Genomics : An Overview of Tools and Insights Into The Genus Corynebacterium. J. Bacteriol. Parasitol. 4(2):1.
Crossref
|
|
|
|
Aritua V, Parkinson N, Thwaites R, Heeney JV, Jones DR, Tushemereirwe W, Smith J (2008). Characterization of the Xanthomonas sp . causing wilt of enset and banana and its proposed reclassification as a strain of X . vasicola. Plant Pathol. 57:170-177.
|
|
|
|
Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS (2011). Dynamic Evolution of Pathogenicity Revealed by Sequencing and Comparative Genomics of 19 Pseudomonas syringae Isolates. PLoS Pathog. 7(7):e1002132.
Crossref
|
|
|
|
Buttner D, He SY (2009). Type III Protein Secretion in Plant Pathogenic Bacteria. Plant Physiol. 150:1656-1664.
Crossref
|
|
|
|
Casabuono A, Petrocelli S, Ottado J, Orellano EG, Couto AS (2011). Structural Analysis and Involvement in Plant Innate Immunity of Xanthomonas axonopodis pv. citri Lipopolysaccharide. J. Biol. Chem. 286:25628-25643.
Crossref
|
|
|
|
Coburn B, Sekirov I, Finlay BB (2007). Type III secretion systems and disease. Clin. Microbiol. Rev. 20(4):535-549.
Crossref
|
|
|
|
Costa TRD, Felisberto-rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G (2015). Secretion systems in Gram-negative insights. Nat. Pub. Group 13(6):343-359.
|
|
|
|
da Silva ACR, Ferro JA, Reinach FC, Farah CS, Furlan LR, Quaggio RB, Monteiro-Vitorello CB, Van Sluys MA, Almeida NF, Alves LMC, do Amaral AM, Bertolini MC, Camargo LEA, Camarotte G, Cannavan F, Cardozo J, Chambergo F, Ciapina LP, Cicarelli RMB, Coutinho LL, Cursino-Santos JR, El-Dorry H, Faria JB, Ferreira AJS, Ferreira RCC, Ferro MIT, Formighieri EF, Franco MC, Greggio CC, Gruber A, Katsuyama AM, Kishi LT, Leite RP, Lemos EGM, Lemos MVF, Locali EC, Machado MA, Madeira AMBN, Martinez-Rossi NM, Martins EC, Meidanis J, Menck CFM, Miyaki CY, Moon DH, Moreira LM, Novo MTM, Okura VK, Oliveira MC, Oliveira VR, Pereira HA, Rossi A, Sena JAD, Silva C, de Souza RF, Spinola LAF, Takita M A, Tamura RE, Teixeira EC, Tezza RID, Trindade dos Santos M, Truffi D, Tsai SM, White FF, Setubal JC, Kitajima JP (2002). Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417(6887):459-463.
Crossref
|
|
|
|
Desaki Y, Miya A, Venkatesh B, Tsuyumu S, Yamane H, Kaku H, Minami E, Shibuya N (2006). Bacterial Lipopolysaccharides Induce Defense Responses Associated with Programmed Cell Death in Rice Cells. Pla. Cell Physiol. 47(11):1530-1540.
Crossref
|
|
|
|
Dunger G, Guzzo CR, Andrade MO, Jones JB, Farah CS (2014). Xanthomonas citri subsp. citri Type IV Pilus Is Required for Twitching Motility, Biofilm Development, and Adherence. Mol. Plant Microbe Interact. 27(10):1132-1147.
Crossref
|
|
|
|
Giltner CL, Nguyen Y, Burrows LL (2012). Type IV pilin proteins: Versatile molecular modules. Microbiol. Mol. Biol. Rev. 76(4):740-772.
Crossref
|
|
|
|
Jalan N, Kumar D, Andrade MO Yu F, Jones JB, Graham JH., White FF, Setubal JC, Wang N (2013). Comparative genomic and transcriptome analyses of pathotypes of Xanthomonas citri subsp . citri provide insights into mechanisms of bacterial virulence and host range. BMC Genomics 14(1):551.
Crossref
|
|
|
|
Koressaar T, Remm M (2007). Enhancements and modifications of primer design program Primer3. Bioinformatics 23(10):1289-1291.
Crossref
|
|
|
|
Kubiriba J, Karamura EB, Tushemereirwe WK, Tinzaara W (2012). Community mobilization : A key to effective control of banana xanthomonas wilt. J. Dev. Agric. Econ. 4(5):125-131.
Crossref
|
|
|
|
Kvitko BH, Park DH, Velásquez AC, Wei CF, Russell AB, Martin GB, Schneider DJ, Collmer A (2009). Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5(4):e1000388.
Crossref
|
|
|
|
Mahuku GS (2004). A simple extraction method suitable for PCR ¬ based analysis of plant, fungal, and bacterial DNA. Plant Mol. Biol. Rep. 22(1):71-81.
Crossref
|
|
|
|
Mhedbi-Hajri N, Darrasse A, Pigné S, Durand K, Fouteau S, Barbe V, Manceau C, Lemaire C, Jacques MA (2011). Sensing and adhesion are adaptive functions in the plant pathogenic xanthomonads. BMC Evol. Biol. 11(1):67.
Crossref
|
|
|
|
Mohammed A (2015). Importance and Characterization of Coffee Berry Disease (Colletotrichum kahawae) in Borena and Guji Zones, Southern Ethiopia. J. Plant Pathol. Microbiol. 6(9):1-6.
Crossref
|
|
|
|
Munford RS, Varley AW (2006). Shield as Signal: Lipopolysaccharides and the Evolution of Immunity to Gram-Negative Bacteria. PLoS Pathog. 2(6):e67.
Crossref
|
|
|
|
Nam J (2001). New Aspects of Gene-for-Gene Interactions for Disease Resistance in Plant. J. Plant Pathol. Microbiol. 17(2):83-87.
|
|
|
|
Newman MA, von Roepenack E, Daniels M, Dow M (2000). Lipopolysaccharides and plant responses to phytopathogenic bacteria. Mol. Plant Pathol. 1(1):25-31.
Crossref
|
|
|
|
Orth K, Xu Z, Mudgett MB, Bao ZQ, Palmer LE, Bliska JB, Mangel WF, Peeters N, Lechner E, Vailleau F, Baud C, Gentzbittel L, Sartorel E, Genschik P, Boucher C (2006). Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. PNAS 103:14620-14625.
Crossref
|
|
|
|
Orth K, Xu Z, Mudgett M B, Bao Z Q, Palmer E L, Bliska J B, Mangel F W, Staskawicz B, Dixon E J. (2000). Homologs Disruption of Signaling by Yersinia Effector YopJ, a Ubiquitin-Like Protein Protease. Science, 290;1594-1597.
Crossref
|
|
|
|
Pizarro-Cerda J, Cossart P (2006). Bacterial Adhesion and Entry into Host Cells. Cell 124:715-727.
Crossref
|
|
|
|
Qian W, Jia Y, Ren S, He Y, Feng J, Lu L, Sun Q, Ying G, Tang D, Jie T, Hua W, Wei H, Pei W, Lifeng J, Bo L, Zeng S, Gu W, Yi L, Gang R, Li T, Yingchuan Y, Zhijian F, Gang C, Baoshan F, Rongxiang Q, Boqin C, Zhu Z, Guo PT, Ji L, He C (2005). Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv . campestris. Genome Res. 15(6):757-767.
Crossref
|
|
|
|
Sluys MA, Camargo LEA, Menck CFM, Silva ACR, Ferro JA, Oliveira MC, Setubal JC, Kitajima JP, Simpson AJ (2002). Comparative Genomic Analysis of Plant Associated Bacteria. Ann. Rev. Phytopathol. 40:169-189.
Crossref
|
|
|
|
Ssekiwoko F, Tushemereirwe WK, Batte M, Ragama P, Komakech A (2006). Reaction of Banana Germplasm to innoculation with Xanthomonas campestris pv. musacearum. Afr. Crop Sci. J. 14(2):151-155.
|
|
|
|
Studholme DJ, Kemen E, Maclean D, Schornack S, Aritua V, Thwaites R, Grant M, Smith J, Jones JDG (2010). Genome-wide sequencing data reveals virulence factors implicated in banana Xanthomonas wilt. FEMS Microbiol. Lett. 310:182-192.
Crossref
|
|
|
|
Studholme DJ, Wasukira A, Paszkiewicz K, Aritua V (2011). Draft Genome Sequences of Xanthomonas sacchari and Two Banana-Associated Xanthomonads Reveal Insights into the Xanthomonas Group 1 Clade. Genes 2(4):1050-1065.
Crossref
|
|
|
|
Todar K (2014). Colonization and Invasion by Bacterial Pathogens. Text Book of Bacteriology. pp. 3-5.
|
|
|
|
Tripathi L, Mwaka H, Tripathi JN, Tushemereirwe WK (2010). Expression of sweet pepper Hrap gene in banana enhances resistance to Xanthomonas campestris pv. musacearum. Mol. Plant Pathol. 11(6):721-731.
Crossref
|
|
|
|
Tripathi L, Tripathi JN (2009). Relative susceptibility of banana cultivars to Xanthomonas campestris pv. musacearum. Afr. J. Biotechnol. 8(20):5343-5350.
|
|
|
|
Tushemereirwe WK, Kangire A, Kubiriba J, Maureen N, Gold SC (2004). Diseases threatening banana biodiversity in Uganda. Afr. Crop. Sci. J. 12(1):19-26.
Crossref
|
|
|
|
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012). Primer3-new capabilities and interfaces. Nucleic Acids Res. 40(15):1-12.
Crossref
|
|
|
|
Volk W (1966). Cell wall lipopolysaccharides from xanthomonas species. J. Bacteriol. 91(1):39-42.
|
|
|
|
Wasukira A, Coulter M, Al-Sowayeh N, Thwaites R, Paszkiewicz K, Kubiriba J, Smith J, Grant M, Studholme DJ (2014). Genome Sequencing of Xanthomonas vasicola pv. vasculorum Reveals Variation in Plasmids and Genes Encoding Lipopolysaccharide Synthesis, Type-IV Pilus and Type-III Secretion Effectors. Pathogens 3(1):211-237.
Crossref
|
|
|
|
Wasukira A, Tayebwa J, Thwaites R, Paszkiewicz K, Aritua V, Kubiriba J, Smith J, Grant M, Studholme DJ (2012). Genome-wide sequencing reveals two major sub-lineages in the genetically monomorphic pathogen Xanthomonas campestris pv. musacearum. Genes 3(3):361-377.
Crossref
|
|
|
|
White FF, Chittoor JM, Leach JE, Young SA, Zhu W (1996). Molecular analysis of the interaction between Xanthomonas oryzae pv . oryzae and rice. In Rice genetics III. Proceedings of the Third International Rice Genetics Symposium, 16-20 Oct 1995. Manila (Philippines): IRRI. pp. 255-266.
|