ABSTRACT
High quality RNA extraction from field-grown jute plants can be difficult due to the presence of complex polyphenolic, polysaccharide and waxes. But isolation of RNA in high quality is a basic need in plant genetics, genomics, transcriptomics, molecular biology and related physiological investigations. Here, cetyl trimethyl ammonium bromide (CTAB) based modified RNA isolation protocol suitable for isolating high quality total RNA from different parts of field-grown jute plants was reported. Modifications come with two extra wash with extraction buffer, residual polysaccharides precipitation with absolute ethanol and potassium acetate during phenol:chloroform:isoamyl alcohol (PCI), intermediate pelleting with lithium chloride followed by dissolving the pellet in sodium dodecyl sulfate (SDS). The 28S/18S ratio and RIN of isolated RNA were greater than 2.0 and 7.3 to 8.8, respectively reveal RNA to be high purity. The isolated RNA can be used directly for subsequent downstream applications including cDNA library construction, reverse transcription polymerase chain reaction (RT-PCR) and RNA-Seq raw data generation without further purification. Moreover, this study showed that this method could also be successfully applied to other polyphenols and polysaccharides rich malvaceous plants.
Key words: RNA isolation, Corchorus, Jute, quantitative polymerase chain reaction with reverse transcription (RT-qPCR), cDNA library construction, next-generation sequencing (NGS) data generation.
cDNA sequencing (RNA-Seq) technology provides the opportunity to identify all expressed transcripts (Guttman et al., 2010) along with the assessment of different important phenomena such as gene expression levels (Pickrell et al., 2010), differential splicing events (Liu et al., 2012; Wang et al., 2008, 2010) and allele-specific gene expression (Rozowsky et al., 2011). High quality RNA extraction is required for cDNA sequencing which is very challenging for the plants rich in mucilage, polysaccharides and polyphenolic compounds (Mac, 2007). Jute (Corchorus species) is an annual, herbaceous bast fibre crop and the second most important natural
fiber after cotton, in terms of production and usage. Fibers are commonly used as filler, or reinforcement; insulation or use as structural elements, and disposable or durable products such as yarns and textiles, ropes, twines, non-woven fabrics, paper and fiber board products, packaging and construction materials, geotextiles and composites and automotive parts (van Dam and Bos, 2004). Jute plant has extremely high contents of acidic and proteinaceous type of mucilage (Stephen et al., 2006) in its various tissues which complicates the isolation of RNA for downstream applications (Rai et al., 2010; Pandey et al., 1996). Elimination of this mucilage is a major challenge for the available protocols of RNA extraction. Moreover, the problem is magnified by conformational changes in mature jute leaves. An irreversible increase in viscosity of the hydrocolloid obtained from leaves occurs during cell lysis at the temperature >60°C (Yamazaki et al., 2009). Standard RNA isolation techniques such as guanidinium-phenol-chloroform extraction (Chomczynski and Sacchi, 1987), TRIZOL and Cetyl Trimethyl Ammonium Bromide (CTAB) methods have failed to isolate adequate and quality RNA from jute leaf (Figure S1 and Table 2). The difficulties of RNA extraction from jute plants have been reported by number of researchers (Khan et al., 2004; Mahmood et al., 2011).
Several protocols (Kumar et al., 2011; Sharma et al., 2003; Hu et al., 2002) are available for isolating RNA from plant tissues containing high levels of polysaccharides and polyphenol compounds. Each of the methods is designed for specific plant species and the nature of the target tissue. Samanta et al. (2011) had established a method for RNA extraction from jute stem only, which employs modified hot borate treatment followed by isopycnic centrifugation. Furthermore, Choudhary et al. (2016) also claimed an RNA isolation procedure from developing jute stem only. But the amount of mucilage, polysaccharides and polyphenolic compounds varies extensively within the developing tissues (Khanuja et al., 1999). Therefore, an efficient protocol is required for isolation of quality RNA from all parts of full grown jute plant. Here, a robust plant RNA isolation protocol, suitable for different types of tissue of jute ranging from root to developing fruits, derived from CTAB method was described (Doyle and Doyle, 1987).
Plant sample
Plants of the tossa jute (Corchorus olitorius variety O-4) were grown in an experimental field at Jute Agricultural Experimental Station, Manikganj, Bangladesh. The root, stem, bark, stick and leaf tissues were sampled from 45 days plants. Flowers were collected just after blooming and developing fruits were collected at 15 days post-anthesis. All samples were snap-frozen in liquid N2 and stored at -80°C. To check the general effectiveness of our protocol, RNA was extracted from the leaves of some other malvaceous plants including Hibiscus cannabinnas, Hibiscus sabdariffa, Hibiscus rosa-sinensis, and Abelmoschus esculentus collected from Jute Agricultural Experimental Station, Manikganj, Bangladesh.
Reagents and solutions
Extraction buffer 100 mM Tris-HCl [Invitrogen, USA], pH 8.0; 1.4 M NaCl [Carl Roth, Germany]; 20 mM EDTA [Sigma-Aldrich, Germany], pH 8.0; 2% (w/v) CTAB [Sigma-Aldrich, Germany]; 4% (w/v) Polyvinylpolypyrrolidone (PVPP) [Sigma-Aldrich, Germany]; 2% (w/v) Polyvinylpyrrolidone (PVP) [Sigma-Aldrich, Germany]; 4% (v/v) β-mercaptoethanol [Sigma-Aldrich, Germany] (to be added just before use); Phenol [Carl Roth, Germany]:Chloroform [Sigma-Aldrich, Germany]:isoamyl alcohol [Carl Roth, Germany] (PCI; 25:24:1, v/v; freshly prepared); 10 M LiCl [Sigma-Aldrich, Germany]; 2% SDS [Invitrogen, USA]; 5 M Potassium acetate [Sigma-Aldrich, Germany], pH 4.8; Absolute Ethanol [Merck, Germany]; Isopropanol [Sigma-Aldrich, Germany]; 70% ethanol. The chemicals used were of molecular biology grade and solutions were made with 0.1% v/v diethylpyrocarbonate (DEPC) [Carl Roth, Germany] treated water and consumables were RNase-free as well.
Isolation procedure
One gram frozen tissue of each sample was ground to a very fine powder in liquid nitrogen with a cooled mortar and pestle. The powder was transferred immediately (before thawing) to 20 ml Extraction Buffer (EB) kept in a 50 ml falcon tube which was pre-heated at 65°C in water bath. The sample in extraction buffer was homogenized by vigorous shaking followed by incubation at 65°C for 20 min with occasional shaking. After incubation, the homogenate was centrifuged at 13,000 g for 20 min at room temperature. The supernatant was collected carefully without disturbing the pellet and transferred to 50 ml tube containing 10 ml EB buffer and inverted to mix the contents. After 10 min incubation at room temperature, the content was centrifuged at 13,000 g for 20 min at room temperature and supernatant was collected carefully without disturbing the pellet in 50 ml tube containing 5 ml EB buffer and inverted to mix the contents followed by 10 min incubation at room temperature. Again the content was centrifuged at 13,000 g for 20 min at room temperature. This time the supernatant was collected and kept on ice until further steps. One tenth volume of absolute ethanol and 1/15 volume potassium acetate (5 M, pH 4.8) were added to the tube and mixed by inversion. Then an equal volume of PCI (25:24:1, v/v) was added, shaked vigorously by hand for 10 s and the content was turned into milky white in colour. The mixer was centrifuged at 13,000 g for 20 min at 4°C and the aqueous phase was collected in a 15 ml falcon tube kept on ice. And then, 1/4 volume of 10 M LiC was added, mixed gently, incubated for 1 h at -70°C (at this stage, sample can be stored at -20°C if it is required to perform the rest of the procedure later) and centrifuged at 13,000 g for 20 min at 4°C. The supernatant was discarded and the pellet was dissolved in 1 ml SDS (2%, w/v). An equal volume of PCI (25:24:1, v/v) was added and mixed well by inversion followed by centrifugation at 13,000 g for 20 min at 4°C. The resulting upper aqueous phase was transferred to a 1.5 ml Eppendorf tube. Then, an equal volume of ice-cold 2-propanol was added and mixed well (at this point 2-propanol added sample can be stored at -20°C if it is required to perform the rest of the procedure later). After incubation for 1 h at -70°C, the content was centrifuged at 13,000 g for 20 min at 4°C and the supernatant was discarded. The final RNA pellet was washed twice with 70% ethanol by centrifugation at 13,000 g for 10 min at 4°C. After confirmation of complete removal of ethanol by air-drying, the pellet was dissolved in nuclease free water.
This isolation procedure basically differs from traditional CTAB based method in such ways: two extra wash with EB buffer, adding absolute ethanol and potassium acetate during first phenol: chloroform:isoamyl alcohol (PCI) step, first precipitation with LiCl followed by dissolving first pellet with SDS. All those modifications were done to reduce the amount of mucilage, polysaccharides and polyphenolic compounds present in different parts of jute plant as well as to ensure the purity of RNA.
Assessment of RNA quantity and integrity
The RNA quantity and purity were checked both spectro-photometrically using NanoDrop® (ND2000c, Thermo Scientific, USA) and by electrophoresis followed by visualization on a denaturing formaldehyde 1.2% agarose gel using Molecular Imager® Gel DOCTM XR+ with Image LabTM Software (Bio Rad Laboratories, Inc., USA). Quality and integrity of isolated RNA was also verified electrophoretically on an Agilent 2100 Bioanalyzer with RNA 6000 Nano LabChip (Agilent Technologies, CA, USA).
cDNA synthesis and RT-PCR analysis
RNA extracted from the samples was treated with DNase I amplification grade (Sigma-Aldrich, Germany) to remove any traces of genomic DNA according to the manufacturer’s instructions. cDNA was synthesized from 1.0 µg of total RNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc) containing oligo dT primers according to manufacturer’s instructions. Freshly prepared cDNAs were amplified by GAPDH gene specific primers (Forward primer 5'- GAAGGATCGGTAGGTTGGTG -3' and Reverse primer 5'-CCTTGACTTTGAGCTCGTGA -3'). The primer was designed using Genscript primer designing tool with default parameters. PCR reactions were carried out in a final volume of 25 µl reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 µM dNTP, 0.2 µM each primer, 2.0 µl cDNA and 0.5 U of Taq DNA polymerase. PCR was performed using thermo cycler condition which was initiated by a hot start at 95°C for 5 min followed by 30 cycles of 95°C for 30 s, 50°C for 30 s and 72°C for 45 s with a 72°C for 5 min final extension. Minus RT control PCR was carried out with 0.5 µg of total RNA instead of cDNA to check DNA contamination. In addition, a H2O control (without template) was set to check whether the PCR signal is resulting from specific cDNA amplification. The PCR products were resolved on a 1.5% agarose gel and visualized under ultraviolet light.
Quantitative polymerase chain reaction with reverse transcription (RT-qPCR)
RT-qPCR amplification was performed in a LightCycler®480 Instrument II (Roche) using the SYBR Green I Master (Roche) according to manufacturer’s instructions. PCR amplification was carried out in a 20-μl volume containing 10-μl of 2X SYBR Green I Master mix, 10 ng cDNA and 0.4 μM of each primer. Thermo cycling conditions were set as an enzyme activation step at 95°C for 5 min followed by 40 cycles consisting of 10 s of denaturation at 95°C, 10 s of annealing at 50°C and 20 s of elongation at 72°C.
NGS data generation from isolated RNA
Sequencing library was prepared using TruSeq Stranded mRNA Library Prep Kit (Illumina Inc., USA) according to manufacturer’s instructions. mRNA was isolated from 1 μg of total RNA using poly-T oligo attached magnetic beads with two rounds of purification. The purified mRNA was fragmented and primed with random hexamers. First strand cDNA was synthesized using SuperScript II Reverse Transcriptase followed by the synthesis of second-strand cDNA. After end repair and the ligation of adaptors, the products were enriched and amplified by PCR. The amplified cDNA libraries were purified by Agencourt AMPure XP beads and assessed on an Agilent Bioanalyzer 2100 system. After validation and quantification, the final cDNA libraries were sequenced using an Illumina HiSeq™ 2500 sequencing platform.
Bioinformatic analysis
After the removal of the adapters (by Trimmomatic tool version 0.36 with default parameters) from the raw reads, low quality and duplicate reads were filtered using SICLE (Q30+; Min Len=50 bp). HISAT2 2.1.0 was used to map the filtered reads with reference genome assembly and coding sequences of C. olitorius (Wang et al., 2008) with the parameters ‘--mp 4,1 --sp 1,0.5 --score-min L,0,-0.8’.
Data availability
The RNA-Seq data were deposited in the NCBI Sequence Read Archive (SRA) under SRP049494.
Quality, integrity and quantity of extracted RNA
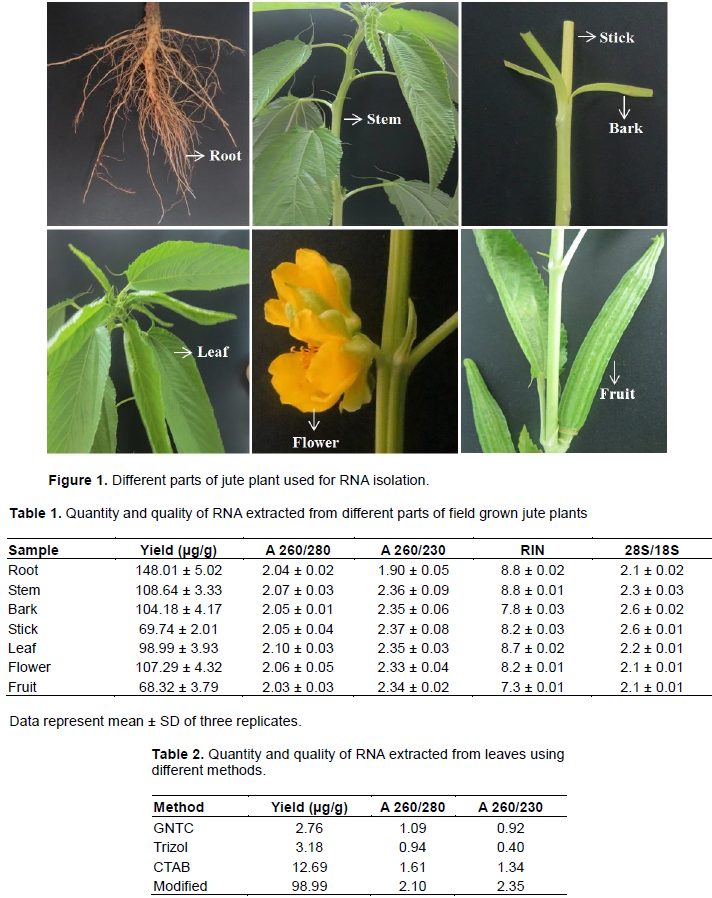
RNA was isolated from various parts of fully-grown jute plant such as root, stem, bark, stick, leaf, flower and fruit (Figure 1) using the CTAB based RNA extraction protocol. The amount of total extracted RNA varied in different parts ranging from 68.32 to 148.01 μg/g tissue (Table 1). Different amount of water and mucilage in different parts of the plant may cause the variation.
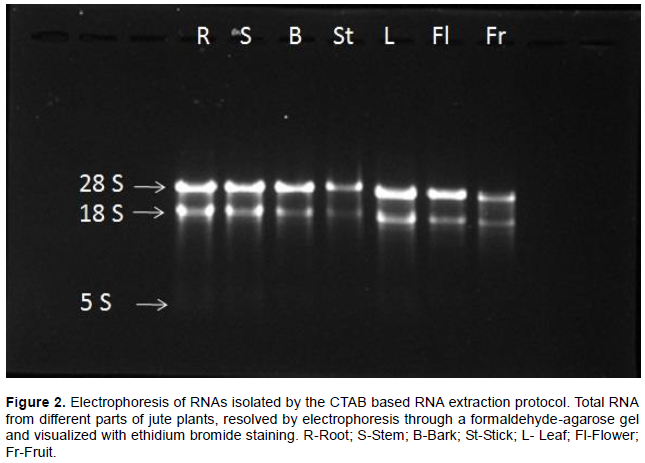
The quality of isolated RNA was assessed by multiple independent methods. The 1.2% agarose gel electro-phoresis (Figure 2) showed that the brightness of 28S rRNA was significantly prominent than 18S rRNA. It demonstrates that the extracted RNAs were not degraded and there was no detectable amount of gDNA contamination. In addition, from the Bioanalyzer analysis, it was observed that the ratio of 28S rRNA and 18S rRNA for different samples were between 2.1 and 2.6 which indicates good quality of RNA (Table 1).
The spectrophotometer analysis showed that the A260/280 and A260/230 absorbance ratio of RNA samples are >2.0 and between 1.90 and 2.37, respectively (Table 1). The A260/280 absorbance ratios represent the high purity of isolated RNAs whereas the A260/230 absorbance ratios indicate that RNAs were not contaminated by polyphenolics and polysaccharides. Though A260/230 absorbance ratio of root little bit lower but the other quality parameters such as RNA gel electrophoresis, yield per g tissue and RIN in bioanalyzer analysis were good. RNA extracted with modified method produced better results compared to CTAB, Trizol and GNTC according to spectrophotometrical and agarose gel electrophoresis analysis (Table 2 and Figure S1).
The RNA integrity was measured using Bioanalyzer metrics (Agilent Technologies, Santa Clara, California, USA). The obtained RNA Integrity Numbers (RIN) were between 7.3 and 8.8 (Table 1) whereas RNA with a RIN value above 7.0 is considered good enough for next-generation sequencing (Islam et al., 2017). A representative electropherogram of RNA isolated from different parts of jute plant are as shown in Figure 3 with clear peaks of rRNAs, indicating no degradation of the RNAs.
Downstream analysis results
RT-PCR
Single-stranded cDNA was successfully synthesized from extracted RNA samples using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc.) containing oligo dT primer. Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) gene was amplified from synthesized cDNA by PCR (Figure 4). Fragments for
GAPDH were obtained as per designed primer (159 bp) in all of the samples, indicating that the isolated RNA is good enough for RT-PCR. No band associated with genomic DNA contamination was observed when RNA was used as template.
RT-qPCR
qRT-PCR was successfully performed with housekeeping gene, GAPDH for further evaluation of the integrity of extracted RNA. A dissociation curve with single peak ensured that the primers amplified specific fragment (Figure 5). In addition, PCR efficiency of GAPDH was determined from the standard curve based on serial dilution. The efficiency values were from 92.72 to 99.04% with good linear regression (R2>0.99) which indicates that there were no PCR inhibitors present in the RNA samples (Figure S2).
Each PCR reaction included minus RT and H2O (no template) control to check for potential genomic DNA and reagent contamination. All samples were amplified in technical and biological triplicates.
NGS data generation
A total of ~53 Gbp RNA-seq data were generated from three independent biological replica samples of jute stem using Illumina HiSeq 2500. About 94 and 83% filtered reads were mapped to reference genome and coding sequences, respectively. Among 37,031 predicted genes in C. olitorius genome (Islam et al., 2017), 27,500 genes were found in RNA-seq data (Supplementary Table 1).
Applicability in malvaceous plant species other than jute
The described protocol for RNA isolation can also be applied for various mucilage rich plant species. This protocol was used for four malvaceous plant species, H. cannabinnas, H. sabdariffa, H. rosa-sinensis, and A. esculentus and found excellent results (Figure S3 and Supplementary Table 2).
Trizol, GNTC protocol and CTAB method were initially tried for RNA extraction from different parts of fully-grown jute plant. Due to presence of high polysaccharide and other viscous materials in the samples, both Trizol and GNTC protocol failed to collect desired supernatant from phase separation step resulting RNA recovery cumbersome. RNA extracted with modified method produced better results compared to CTAB, Trizol and GNTC according to spectrophotometrical analysis (Table 2). Similarly, electrophoresis in agarose gel shows that the quality and quantity of RNA obtained from CTAB, Trizol and GNTC method is not adequate enough (Figure S1).
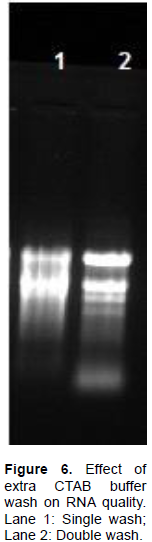
Therefore, the CTAB method was modified. A high-strength CTAB solution was used as a lysis buffer with a higher concentration of PVPP, PVP and β-mercaptoethanol. It ensured rapid removal of phenolic compounds, which otherwise oxidize and bind to RNA molecules. The combination of PVP and PVPP produced top aqueous phase immediately displayed transparency, indicating successful elimination of pigments and most of polyphenolics. In order to reduce the amount of mucilage, two additional CTAB buffer washes were included in our protocol which could successfully eliminate pigments and most polyphenolics (Figure 6). Further, in first purification step of this protocol, 1/10 V absolute ethanol and 1/15 V potassium acetate were added to the supernatant along with phenol: chloroform: isoamyl alcohol (PCI). Absolute ethanol and KAc effectively precipitated residual polysaccharides (Yockteng et al., 2013; Yang et al., 2008). LiCl was used in the first precipitation step of the protocol to reduce co-precipitation of polysaccharides with RNA. It is an important step as LiCl does not precipitate DNA, proteins and phenolic compounds. In addition, 2% SDS was used to dissolve precipitated RNA followed by second PCI extraction as SDS effectively disrupts protein-nucleic acid interaction and protect RNA from ribonucleases. After this disruption, protein was denatured by PCI and became insoluble in aqueous solution (Rio et al., 2010), resulting in good quality RNA with no protein contaminants (Table 1).
There are a limited number of published methods to isolate RNA from jute plant. For instance, Mahmood et al. (2011) reported the procedure to isolate RNA from jute seedling and both Samanta et al. (2011) and Choudhary et al. (2016) illustrated RNA isolation methods from jute stem only. However, the protocol, claimed by Samanta et al. (2011), requires ultra-centrifuge facility, which is a very complex and expensive setup and might not be available in regular laboratories, and costly reagent CsCl for purification of RNA. In contrast, the procedure described here is highly reproducible and easily adoptable in usual laboratory conditions and suitable for isolating RNA from plant species and different tissues rich in polysaccharides and phenolic compounds. To our knowledge, this is the first report of a single method which can be utilized to isolate high quality RNA from different parts of field grown jute plant. Furthermore, using this protocol, high quality RNA has been extracted from different plant species in Malvaceae (H. cannabinnas, H. sabdariffa, H. rosasinensis, and A. esculentus) containing abundant polyphenols and polysaccharides.
This method is highly reproducible and easily adoptable in usual laboratory conditions and suitable for isolating RNA from different developing parts of jute plant. The RNA obtained by using this method can be used directly for subsequent downstream applications including cDNA library construction, qRT-PCR and RNA-seq data generation without further purification. The method also applied to other polyphenols and polysaccharides rich malvaceous plant.
The authors thank Md Sharifur Rahman (Department of Telecommunications, Dhaka 1208, Bangladesh) for critical suggestions and comments on the manuscript. They are grateful to Emdadul Mannan Emdad (Basic and Applied Research on Jute Project, Bangladesh Jute Research Institute, Dhaka 1207, Bangladesh) for technical help. This research was funded by the Government of Bangladesh.
The authors have not declared any conflict of interests.
REFERENCES
|
Chomczynski P, Sacchi N (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Analytical Biochemistry 162:156-159.
Crossref
|
|
|
|
Choudhary SB, Kumar M, Chowdhury I, Singh RK, Pandey SP, Sharma HK, Karmakar PG (2016). An efficient and cost effective method of RNA extraction from mucilage, phenol and secondary metabolite rich bark tissue of tossa jute (C. olitorius L.) actively developing phloem fiber. 3 Biotech 6:100.
Crossref
|
|
|
|
|
Doyle JJ, Doyle JL (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19:11-15.
|
|
|
|
|
Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A (2010). Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nature Biotechnology 28:503-510.
Crossref
|
|
|
|
|
Hu CG, Honda C, Kita M, Zhang Z, Tsuda T, Moriguchi T (2002). A simple protocol for RNA isolation from fruit trees containing high levels of polysaccharides and polyphenol compounds. Plant Molecular Biology Report 20:69a-69g.
Crossref
|
|
|
|
|
Islam MS, Saito JA, Emdad EM, Ahmed B, Islam MM, Halim A, Hossen QMM, Hossain MZ, Ahmed R, Hossain MS, Kabir SM, Khan MS, Khan MM, Hasan R, Aktar N, Honi U, Islam R, Rashid MM, Wan X1, Hou S, Haque T, Azam MS, Moosa MM, Elias SM, Hasan AM, Mahmood N, Shafiuddin M, Shahid S, Shommu NS, Jahan S, Roy S, Chowdhury A, Akhand AI, Nisho GM, Uddin KS, Rabeya T, Hoque SM, Snigdha AR, Mortoza S, Matin SA, Islam MK, Lashkar MZ, Zaman M, Yuryev A, Uddin MK, Rahman MS, Haque MS, Alam MM, Khan H, Alam M (2017). Comparative genomics of two jute species and insight into fibre biogenesis. Nature Plants 3:16223.
Crossref
|
|
|
|
|
Khan F, Islam A, Sathasivan AK (2004). Rapid method for high quality RNA isolation from jute: Corchorus capsularis L. and Corchorus olitorius L. Plant Tissue Culture & Biotechnology 14:63-68.
|
|
|
|
|
Khanuja SPS, Shasany AK, Darokar MP, Kumar S (1999). Rapid isolation of DNA from dry and fresh samples of plants producing large amounts of secondary metabolites and essential oils. Plant Molecular Biology Report 17:74.
Crossref
|
|
|
|
|
Kumar GRK, Eswaran N, Johnson TS (2011). Isolation of high-quality RNA from various tissues of Jatropha curcas for downstream applications. Analytical Biochemistry 413:63-65.
Crossref
|
|
|
|
|
Liu J, Lee W, Jiang Z, Chen Z, Jhunjhunwala S, Haverty PM, Gnad F, Guan Y, Gilbert HN, Stinson J, Klijn C, Guillory J, Bhatt D, Vartanian S, Walter K, Chan J, Holcomb T, Dijkgraaf P, Johnson S, Koeman J, Minna JD, Gazdar AF, Stern HM, Hoeflich KP, Wu TD, Settleman J, de Sauvage FJ, Gentleman RC, Neve RM, Stokoe D, Modrusan Z, Seshagiri S, Shames DS, Zhang Z (2012). Genome and transcriptome sequencing of lung cancers reveal diverse mutational and splicing events. Genome Research 22:2315-2327.
Crossref
|
|
|
|
|
Mac Rae E (2007). Extraction of plant RNA. In 'Methods in molecular biology, Protocols for nucleic acid analysis by nonradioactive probes'. (Eds Hilario E, Mackay J, 2nd ed.) (New Jersey: Humana Press) 353:15-24.
|
|
|
|
|
Mahmood N, Ahmed R, Azam MS, Khan H (2011). A simple and swift method for isolating high quality RNA from jute (Corchorus spp.). Plant Tissue Culture & Biotechnology 21:207â€211.
Crossref
|
|
|
|
|
Pandey RN, Adams RP, Flournoy LE (1996). Inhibition of random amplified polymorphic DNAs (RAPDs) by plant polysaccharides. Plant Molecular Biology Report 14:17-22.
Crossref
|
|
|
|
|
Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y, Pritchard JK (2010). Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464:768-772.
Crossref
|
|
|
|
|
Rai V, Ghosh SJ, Dey N (2010). Isolation of total RNA from hard bamboo tissue rich in polyphenols and polysaccharides for gene expression studies. Electronic Journal of Biotechnology 13:5.
Crossref
|
|
|
|
|
Rio DC, Ares M Jr, Hannon GJ, Nilsen TW (2010). Purification of RNA by SDS Solubilization and Phenol Extraction. Cold Spring Harbor Protocols. doi: 10.1101/pdb.prot5438
Crossref
|
|
|
|
|
Rozowsky J, Abyzov A, Wang J, Alves P, Raha D, Harmanci A, Leng J, Bjornson R, Kong Y, Kitabayashi N, Bhardwaj N, Rubin M, Snyder M, Gerstein M (2011). AlleleSeq: analysis of allele-specific expression and binding in a network framework. Molecular Systems Biology 7:522.
Crossref
|
|
|
|
|
Samanta P, Sadhukhan S, Das S, Joshi A, Sen SK, Basu A (2011). Isolation of RNA from field-grown jute (Corchorus capsularis) plant in different developmental stages for effective downstream molecular analysis. Molecular Biotechnology 49:109-115.
Crossref
|
|
|
|
|
Sharma AD, Gill PK, Singh P (2003). RNA isolation from plant tissues rich in polysaccharides. Analytical Biochemistry 314:319-321.
Crossref
|
|
|
|
|
Stephen AM, phillips GO, williams PA (2006). Food polysaccharides and their applications. 2nd ed. (Boca Raton: CRC Press).
|
|
|
|
|
van Dam JEG, Bos HL ( 2004). The environmental impact of hard fibres and jute in non-textiles industrial application. Consultation of natural fibres. ESC-Fibres consultation No. 04/4. Short version of comprehensive review on the environmental impact of the natural fibres in industrial applications. Agrotechnology and Food Innovations (A & F), Wageningen UR, Wageningen, The Netherlands.
|
|
|
|
|
Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB (2008). Alternative isoform regulation in human tissue transcriptomes. Nature 456:470-476.
Crossref
|
|
|
|
|
Wang X, Wu Z, Zhang X (2010). Isoform Abundance Inference Provides a More Accurate Estimation of Gene Expression Levels in RNA-Seq. Journal of Bioinformatics and Computational Biology 8:177-192.
Crossref
|
|
|
|
|
Yamazaki E, kurita O, Matsumura Y (2009). High viscosity of hydrocolloid from leaves of Corchorus olitorius L. Food Hydrocolloids 23:655-660.
Crossref
|
|
|
|
|
Yang G, Zhou R, Tang T, Shi S (2008). Simple and Efficient Isolation of High-Quality Total RNA from Hibiscus tiliaceus, a Mangrove Associate and Its Relatives. Preparative Biochemistry and Biotechnology 38:257-264.
Crossref
|
|
|
|
|
Yockteng R, Almeida AMR, Yee S, Andre T, Hill C, Specht CD (2013). A Method for Extracting High-Quality RNA from Diverse Plants for Next-Generation Sequencing and Gene Expression Analyses. Applications in Plant Sciences 1:1300070.
Crossref
|
|