ABSTRACT
Conservation of farm animal genetic resources is of fundamental importance for the study of the relationships among breeds. The aim of this study was to evaluate the usefulness of the nuclear fluorescence inter simple sequence repeat (FISSR) markers in order to shed light on the genetic biodiversity of domestic animals. Two modifications of the original technique were made so as to make it more suitable for routine needs. The modified FISSR protocol was tested on different breeds of goat and donkey from Sardinia, a Mediteranean island known for its biodiversity. The two species are affected by different management problems in Sardinia: goats need a traceability of local products from different breeds, whereas donkeys are drastically reduced in number. The primers used were found to be very informative suggesting that the modified FISSR can be successfully applied in studies on different breeds of animal species without expensive experimentations. This method could be of interest in many geographic regions where there are more breeds of the same species with similar morphological features and different genetic pattern. The strongest point of this method is its low cost.
Key words: Fluorescence inter simple sequence repeat (FISSR), biodiversity, genetic variability, goat, donkey.
The nuclear fluorescence inter simple sequence repeat (FISSR) may achieve new insights on the genetic variability of populations of different species, in particular those involved in animal production (Nagaraja et al., 2005). This method has been termed FISSR-PCR by the inventors, who tested its usefulness in plants (a large number of rice varieties) and insects (Bomby xmori). The results clearl yshowed that this method is an important tool for large scale screening of varieties/cultivars and high throughput genotyping in mapping genomes where genomic information is scanty or absent (Nagaraju et al., 2002).
The FISSR technique is less laborious when compared with other ï¬nger-printing methods and produces highly reproducible bands and results with low statistical error, leading to multilocus and highly polymorphic banding patterns. In addition, it does not require prior knowledge of DNA sequences and could be introduced as an effective tool to gain helpful genetic information from wild, reared and natural populations (Leighton, 2002; Luikart et al., 2003; Casu et al., 2006).
In the present study, two changes in the technique presented by Nagaraju et al. (2002) were introduced to make it more applicable to routine needs, reducing the number of primers and changing the kind of label for analysis by sequencer. Such modified technique was tested in goats (Capra hircus) and donkeys (Equus asinus), two species with different management problems (Dossa et al., 2007; Colli et al., 2013). All the samples came from Sardinia, an island rich in animal biodiversity. In the analysis of goat genotype, the finding of high FISSR polymorphism may allow the molecular traceability of the different breeds, and this may be important for the control of local products and constitutes a quick tool for the economic development of the different breeds. Donkeys were used in agriculture until the middle of the last century. From this period, there was a drastic reduction in their number due to agricultural mechanization. Autochthonous donkeys belong to two different breeds in Sardinia: the Sardinian and the Asinara donkeys. The Sardinian donkey shows a grey coat with a black cross on its back and is spread all over Sardinia. In contrast, Asinara donkey is an albino variant originating from Asinara island, north-western Sardinia. Both breeds are numerically reduced and considered in danger of extinction. According to the FAO risk status classification of animal genetic resources (www.fao.org), Sardinian donkey is an “endangered breed” and Asinara donkey is a “critical breed”. Differentiation of the two donkey breeds is commonly performed on the basis of the morphological traits. Consequently, a genetically characterization of these breeds could be very useful in the studies on the relationships among local farms for the estimation of their relative importance and the correct application of preservation programs, in order to conserve the total genetic variance for this species, as made for other species in the past (Allendorf et al., 2001; Bennewitz and Meuwissen, 2005; Safari et al., 2005; Usai et al., 2006; Kefena et al., 2011; Iquebal et al., 2013).
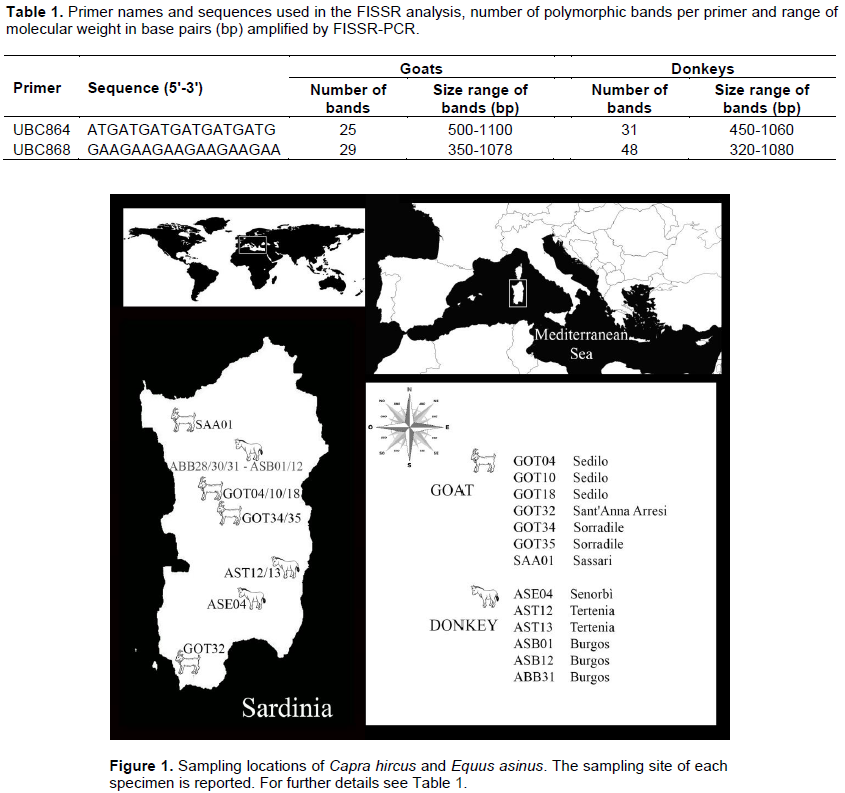
All specimens (seven goats and eight donkeys) and sampling sites are shown in Figure 1. In contrast to the protocol of Nagaraju et al. (2002), where a set of eight degenerate primers (non-dyed) and one fluorescent dye-labeled nucleotide (dUTP TAMARA) were employed, in the present work, only two non-anchored labeled primers were used for FISSR-PCR reaction. A preliminary screening of 29 primers allowed identifying 2 primers that were used to genotype all the individuals: UBC864 and UBC868 (Table 1) labeled respectively with 6-FAM and HEX fluorescent dyes (Sigma-Aldrich, St. Louis, MO, USA). These were used for both goat and donkey. DNA was extracted from 600 μl frozen blood with a standard Salting Out protocol after prewashing for 10 min at room temperature with 600 µl Triton Blood Buffer (0.32 M sucrose, 10 mM Tris HCl, 5 mM MgCl2, 1% Triton X 100). Then, the aliquot was centrifuged at 9,000 g for 8 min, the suspension discarded and the pellet resuspended in a standard lysis buffer suitable for blood extraction. Each 25 μl PCR mixture contained about 100 ng total genomic DNA, PCR buffer, 3.5 mM MgCl2, 0.5 µM primer and 0.2 mM each dNTP, with 0.05 U/μl of Taq DNA polymerase (Sigma-Aldrich). Both positive and negative controls were used to assess the effectiveness of the PCR reagents. The thermal cycling conditions were set as follows: initial denaturation of 3 min at 94° C followed by 35 cycles of: 1 min at 94°C, 2 min at 45°C, 1 min at 72°C and a final extension of 5 min at 72°C. A visual checking was carried out by electrophoresis on 1.5% agarose/TBE gel stained with ethidium bromide (10 mg/ml) at 4 V/cm for 240 min, and all samples were shown to give reproducible band patterns. The samples were genotyped with ROX100 BV labeled molecular weight standard onto an ABI377 automated sequencer, carried out by an external sequencing core service (BMR Genomics, Padua, Italy). Each FISSR electropherogram peak pattern was converted into a binary matrix (1 for band presence and 0 for absence) assuming that each peak represents a single diallelic locus.
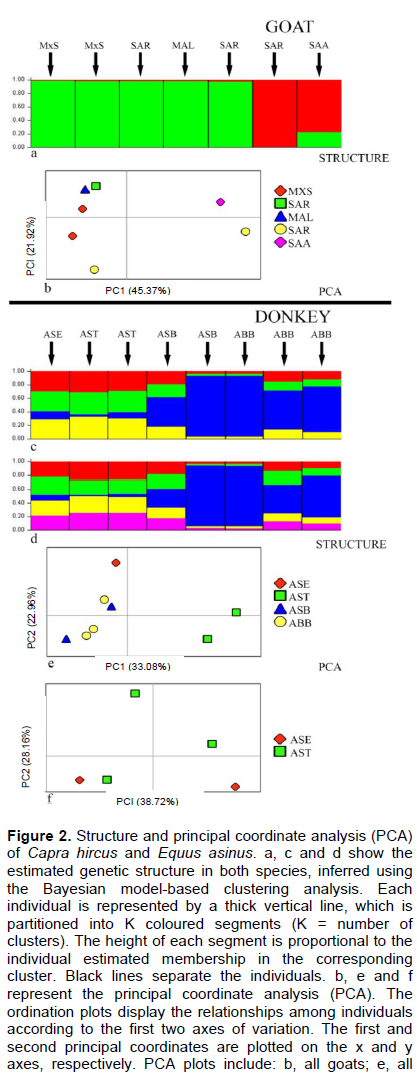
In order to overcome potential problems due to the small sampling plan, the underlying genetic population structure in our preliminary analysis was inferred using an individual-based approach (Luikart et al., 2003) using the Bayesian model-based clustering algorithms implemented in the software structure 2.2.3 (Pritchard et al., 2000). Ten independent analyses were performed to assess the reliability of the results. In each analysis, the number of clusters (K) was estimated by a range of possible values (K=2 up to 8). Structure analysis, which assumes that K is known in advance, was used to assess the occurrence of hierarchical levels of genetic structure. In a given dataset, structure identifies the uppermost hierarchical structure that corresponds to the minimum number of clusters that captures the major structure in the data (Pritchard and Wen, 2004). After this first round, data are partitioned into smaller datasets according to the best clustering solution, and subsequent rounds are performed on each subset of data. The procedure was reiterated until it was not possible to partition the data any further. The results of the model-based clustering was then compared with a principal coordinate analysis (PCA) performed by the program genalex 6.3 (Peakall and Smouse, 2006) on a matrix of inter-individual distances via a covariance matrix with a data standardisation method, in order to avoid artifacts due to violation of the model assumptions (Hardy-Weinberg and linkage equilibrium) or isolation by distance (Guillot et al., 2009). Genetic structuring was not investigated by a hierarchical analysis of molecular variance (AMOVA) because both goats and donkeys had populations of a single individual.
Based on a total of 15 sequences, clearly reproducible patterns were found in the two species that yielded a total of 54 peaks in goats, indicating polymorphic sites, (25 of which for UBC864 and 29 for UBC868) and a total of 79 peaks in donkeys (31 of which for UBC864 and 48 for UBC868). Private bands were detected both in goats and donkeys, and interestingly, the donkey AST12 shows four private bands.
In goats, the size of bands ranged from 500 to 1100 bp for the primer UBC864 and 350 to 1078 bp for the primer UBC868 (Table 1). The uppermost hierarchical level of genetic structuring, as estimated from the first round of structure analyses (Figure 2a), resulted in a clustering solution of K=2. The green cluster corresponded to samples of Sarda (GOT18, GOT34), Maltese (GOT32) and their crossbreeds (GOT04, GOT10), the red cluster corresponded to samples of Saanen (SAA01) and an "alleged" Sarda (GOT35).
The successive round of structure analyses (overall K=4 and K=5, data not shown) did not show a further substructuring, according to the first clustering solution (K=2), which identified two main genetic clusters. The first two principal coordinates resulting from PCA carried out on the entire dataset, accounted for 67.29% of the total
variation, and identified two groups of individuals (Figure 2b). These groups were consistent with the structuring found by structure analyses (Figure 2a). The sample GOT35 from Sorradile (Oristano), was previously morphologically classified as Sarda, grouped with Saanen (SAA01), as it showed a FISSR genetic profile almost perfectly super-imposable, so revealing a probable mistake in taxonomic attribution on morphological bases. Further PCA analysis on a subset of data corresponding to the breeds of Sarda, Maltese and their crossbreeds (data not shown) gave no additional information.
In donkeys, the banding pattern ranged from 450 to 1060 bp for the primer UBC864 and 320 to 1080 bp for the primer UBC868 (Table 1). For this species, the uppermost hierarchical level of genetic structuring resulted in a clustering solution of K=4 (Figure 2c). The genetic structure did not remark the distribution of individuals (despite the numerical evenness) because the two breeds from Burgos (Sardinian and Asinara donkeys) showed one individual (ASB12 and ABB28, respectively) completely characterized by the blue cluster. The successive round of structure analyses (K=5, Figure 2d) showed that the blue cluster roughly corresponding to the samples from Burgos was inconsistent with the samples from Tertenia. Conversely, the donkey from Senorbì had the same probability to belong to one of the five clusters. These results were consistent across replicate runs, which retrieved nearly the same clustering solution. The first two principal coordinates that resulted from PCA carried out on the entire dataset, accounted for 56.03% of the total variation, and identified three groups of individuals (Figure 2e). The four populations are separated according to their geographic origin in three groups, with the sample from Senorbì more distant from those from Tertenia than from Burgos, along the axis 1, which explains most of the variability (33.08%). Further PCA analysis on a subset of data corresponding to the donkeys from Burgos showed that ASB12 and ABB28 grouping is far from the other samples (Figure 2f).
The results shown here demonstrate that modified FISSRs can be successfully used in the studies on population genetics of Capra hircus and Equus asinus and it is conceivable that this technique could be a valid tool in a large number of species, especially those where there are no other genetic markers and not requiring DNA sequencing. In contrast to the classical protocol where a set of degenerate primers (non-dyed) and one fluorescent dye-labeled nucleotide (dUTP TAMARA) were employed, in the present work only two non-anchored labeled primers were used for FISSR-PCR reaction. This method is very useful both in the traceability of animal products and in the management of local breeds that are numerically vulnerable, as is the case of the different breeds belonging to the two species here examined (donkeys and goats). Indeed, both breeds are numerically reduced and considered in danger of extinction. Thus, the genetic characterization of such breeds could be very useful in the studies on the relationships among local farms for the estimation of their relative importance and the correct application of preservation programs. This method can provide a tool for the preservation of local genetic breeds widespread all over the world, giving a contribution to the safeguard of animal biodiversity. Moreover, this method is not expensive as compared to other genetic analyses. It could be of interest in many geographic regions where there are more breeds of the same species with similar morphological features and different genetic patterns. The strongest point of this method is its low cost.
The authors declare that there is no conflict of interest.
REFERENCES
|
Allendorf FW, Leary RF, Spruell P, Wenburg JK (2001). The problems with hybrids: setting conservation Guidelines. Trends Ecol. Evol. 16: 613-622.
Crossref
|
|
|
|
Bennewitz J, Meuwissen THE(2005). Estimation of extinction probabilities of five German cattle breeds by population viability analysis. J. Dairy Sci. 88: 2949-2961.
Crossref
|
|
|
|
Casu M, Casu D, Lai T, Cossu P, Curini-Galletti M (2006). Intersimple sequence repeat markers reveal strong genetic differentiation among populations of the endangered mollusc Patella ferruginea (Gastropoda: Patellidae) from two Sardinian marine protected areas. Mar. Biol. 149:1163-1174.
Crossref
|
|
|
|
Colli L, Perrotta G, Negrini R, Bomba L, Bigi D, Zambonelli P, Verini Supplizi A, Liotta L, Ajmone-Marsan P (2013). Detecting population structure and recent demographic history in endangered livestock breeds: the case of the Italian autochthonous donkeys. Anim. Genet. 44: 69-78.
Crossref
|
|
|
|
Dossa LH, Wollny C, Gauly M (2007). Spatial variation in goat populations from Benin as revealed by multivariate analysis of morphological traits. Small Ruminant Res. 73: 150-159.
Crossref
|
|
|
|
Guillot G, Leblois R, Coulon A, Frantz AC (2009). Statistical methods in spatial genetics. Mol. Ecol. 18: 4734-4756.
Crossref
|
|
|
|
Iquebal MA, Sarika, Dhanda SK, Arora V, Dixit SP, Raghava GP, Rai A, Kumar D (2013). Development of a model webserver for breed identification using microsatellite DNA marker. BMC Genet. 14: 118.
Crossref
|
|
|
|
Kefena E, Beja-Pereira A, Han JL, Haile A, Mohammed YK, Dessie T (2011). Eco-geographical structuring and morphological diversities in Ethiopian donkey populations. Livest. Sci. 141: 232-241.
Crossref
|
|
|
|
Leighton FA (2002). Healts risk assessement of the translocation of wild animals. Rev. Sci. Tech. 21: 187-195.
Crossref
|
|
|
|
Luikart G, England PR, Tallmon D, Jordan S, Taberlet P (2003). The power and promise of population genomics: from genotyping to genome typing. Nat. Rev. Genet. 4: 981-994.
Crossref
|
|
|
|
Nagaraja GM, Mahesh G, Satish V, Madhu M, Muthulakshmi M, Nagaraju J (2005). Genetic mapping of Z chromosome and identification of W chromosome-specific markers in the silkworm, Bombyx mori. Heredity (Edinb.) 95(2): 148-157.
Crossref
|
|
|
|
Nagaraju J, Kathirvel M, Subbaiah EV, Muthulakshmi M, Kumar LD (2002). FISSR-PCR: a simple and sensitive assay for highthroughput genotyping and genetic mapping. Mol. Cell Probe 16: 67-72.
Crossref
|
|
|
|
Peakall R, Smouse PE (2006). Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6: 288-295.
Crossref
|
|
|
|
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945-959.
|
|
|
|
Pritchard JK, Wen W (2004). Documentation for structure software: version 2. University of Chicago Press, Chicago, IL, USA.
|
|
|
|
Safari E, Fogarty NM, Gilmour AR (2005). A review of genetic parameter estimates for wool, growth, meat and reproduction traits in sheep. Livest. Prod. Sci. 92: 271-289.
Crossref
|
|
|
|
Usai MG, Casu S, Molle G, Decandia M, Ligios S, Carta A (2006). Using cluster analysis to characterize the goat farming system in Sardinia. Livest. Sci. 104: 63-76.
Crossref
|