ABSTRACT
The microbial sources used for the production of nanoparticles are important, and one of the reasons for bacterial preference is their ease of manipulation and ecofriendly potential. Three bacteria strains of Streptomyces genus were used for the biosynthesis of silver nanoparticles (AgNPs). The aim of the present study was to confirm Streptomyces spp. as biological systems in the synthesis and extracellular characterization of silver nanoparticles with antimicrobial activity. The nanoparticles were characterized by ultraviolet–visible spectroscopy (UV-Vis), dynamic light scattering (DLS), RDX, Fourier-transform infrared spectroscopy (FTIR) and transmission electron microscope (TEM). In qualitative antimicrobial test, Streptomyces parvulus DPUA 1549 and Streptomyces owasiensis DPUA 1748 showed activity against pathogenic microorganisms (bacteria and yeasts) tested. The AgNPs of S. parvulus DPUA 1549 showed the most efficient minimum inhibitory concentration (MIC) at low concentrations (1.95 μM) against Staphylococcus aureus. They promoted structural damage to the micro-organism cell surface according to AFM images and also reduced the proliferation of NIH-3T3 cells in the cytotoxic assay. Thus, biosynthesis of silver nanoparticles by Streptomyces spp. is a potential source to be used in nanomedicine and cosmetology industries.
Key words: Streptomyces, tropical region, nanobiotechnology, antimicrobial, cytotoxic test.
Actinomycetes are Gram-positive filamentous bacteria present in marine and terrestrial environment. They have an unparalleled ability to produce diverse active compounds such as antibiotics. Over 500 species of actinomycetes have been described, until now, and among them Streptomyces genus provides nearly 80% of the industrially available antibiotics (Abd-Elnaby et al., 2016; Bhosale et al., 2015).
Nanobiotechnology is an innovative basis in the synthesis of nanomaterials and nanostructures by biological sources. Professional researchers such as microbiologists, biochemists, chemists, physicists and other branches of engineering are contributing new solutions and new sources as micro-organisms through approaches of green biochemistry that provide better efficiency and low cost of processes and materials (Buszewski et al., 2016; Gul et al., 2016; Chemat and Strube, 2015).
In the past years, over 15 billion dollars was invested in nanotechnology and there is a projection of at least 3 trillion Euro until 2020. These investments promote advantages in many industrial processes, biotechnology, information technology, agriculture, food industry and health care (Roco et al., 2011; Schneid, 2014). Reports available have shown the potential of actinomycetes as efficient microbial-mediated candidates for synthesis of extracellular and intracellular silver nanoparticles (AgNPs). The development of novel applications in the pharmaceutical field with metallic nanoparticles makes it an attractive alternative to antimicrobial agents. These AgNPs have antimicrobial action against pathogen Gram-positive and Gram-negative bacteria and multi-resistant strains such as methicillin-resistant Staphylococcus aureus (Queiroz et al., 2012; Pantidos and Horsfall, 2014; Durán et al., 2016).
The biosynthesis of nanoparticles by actinomycetes has good stability and polydispersity. They can be genetically manipulated in order to provide better control over the nanoparticles size. In addition, the time required for completion of the reaction using this bacteria ranges between approximately 24 and 120 h (Bhosale et al., 2015). Some species of actinomycetes had shown biological applications of their AgNPs: Streptomyces djkartnsis (Biglari et al., 2014), Streptomyces olivaceus sp-1392 (Evelyne and Subbiayh, 2014), Streptomyces coelicolor (Manikprabhu and Lingappa, 2013), Streptomyces albogriseolus (Samundeeswari et al., 2012), Streoptomyces parvulus SSNP11 and Streptomyces albidoflavus CNP10 (Prakasham et al., 2012) and Streptomyces rochei (Selvakumar et al., 2012).
The understanding of the AgNPs biodistribution/accumulation in living systems is important to make them more effective as pharmaceuticals agents. Researches have demonstrated that, at low concentrations, silver is nontoxic to human cells. It also has been reported that Ag+ ions uncouple the respiratory chain from oxidative phosphorylation or collapse the proton-motive force across the cytoplasmic membrane.
The interaction of Ag+ with bacteria is directly related to the size and shape of the nanoparticles (Manivasagan et al., 2013).
In order to contribute to the development of the biogenic nanoparticles applications, the aim of this research was to evaluate the extracellular microbial-mediated synthesis of silver nanoparticles (AgNPs) by Streptomyces spp. strains, their characterization, antimicrobial and cytotoxic activities.
Microorganisms
Three cultures (DPUA 1549, DPUA 1747 and DPUA 1748) of Streptomyces genus were selected. They were obtained from DPUA Culture Collection - Federal University of Amazonas, Brazil. All of them had already presented antimicrobial activity by liquid fermentation in previous study (Silva-Vinhote et al., 2011). The actinomycetes were reactivated in ISP2A agar (g/L): (starch, 10; yeast extract, 4; malt extract, 10; dextrose, 4; agar, 20), and adjusted to pH 7.3. The cultures were maintained at 30°C for 21 days and authenticated based on the physiologic, macromorphological and micromorphological characteristics of the group (Lechevalier and Lechevalier, 1989; Waskman and Wodruff, 1941).
Identifications of the micro-organisms
The identification was carried out by polymerase chain reaction (PCR). The DNA strain was excreted using the purification kit of genomic DNA (Promega Corporation, Madison, EUA) and the amplification of 16S ribosomal DNA gene was performed by PCR using universal primers from eubacteria ACT235F (5’- CGCGGCCTATCAGCTTGTTG) and ACT878R (5’ - CCGTACTCCCCAGGCGGGG). PCR was conducted in Eppendorf Master Cycle thermocycler. To obtain the final volume of 25 μL, the following were used: 14.2 μL of Milli-Q water, 1.0 μL of denatured cells/DNA, 2.5 μL of DNA polymerase 10× (200 mM Tris-HCl, 500 mM KCl) buffer, 2.5 μL of 25 mM MgCl2, 2.5 μL of 2.5 mM dNTP, 1.0 μL of ACT235F 5’ - 3’5 pmol oligonucleotide, 1.0 μL of ACT878R 5’ - 3’ 5 pmol oligonucleotide and 0.3 μL of 5 U Taq DNA polymerase. The amplification reaction was carried out with the following temperature cycles: (1) 95°C for 5 min, (2) 95°C for 45 s, 72°C for 60 s, 72°C for 60 s (40 times), (3) 72°C for 5 min. The annealing was also made at 68°C. To confirm the amplification, 2.0 μL of the reaction was submitted to electrophoresis in agarose gel with TBE (boric acid-EDTA) and stained with GelRedTM. The electrophoresis was developed in 80 V for 10 min and in 100 V for 50 min. The product was sequenced and the resulting sequence was compared with all sequences available in GenBank using BLAST software of the National Center for Biotechnology Information (NCBI).
Synthesis of silver nanoparticles
Submerged fermentation
The actinomycetes were cultivated in Erlenmeyer flasks (125 mL) containing 50 mL of MPE broth (g/L): (glucose, 20; soybean flour, 20; NaCl, 5; CaCO3, 2). The process was carried out at 25°C, 200 rpm, for 72 h and the biomass was separated from the extract by vacuum using Whatman nº 1 filter. The biomass was filtrated three times with sterilized water and after the last washing; the liquid solution was recovered for silver nanoparticles biosynthesis process.
Silver nanoparticles biosynthesis
Silver nanoparticles synthesis was made in Erlenmeyer flasks (250 ml) containing 50 mL from the recovered solution. A solution of silver nitrate (1 M) was added until final concentration of 1 mM. The reaction was carried out at 25°C, 200 rpm, for 72 h, in the absence of light. The control sample presented only aqueous solution without the silver ion and maintained at same conditions.
Characterization of silver nanoparticles
UV-Visible spectroscopy analysis
The bio-reduction of silver nitrate and the formation of nanostructures were confirmed by UV–Vis spectra (UV-Vis Cary 50 Probe Agilent) in a range of wavelengths between 350 and 900 nm.
Fourier transform infrared (FTIR) spectroscopy analysis
The AgNPs were also examined for the presence of biomolecules responsible for bioreduction using Fourier transform infrared spectral analysis in a Bomem MB Series-B102 Spectrometer. FTIR analysis reveals the biomolecules responsible for the silver ion reduction and the stabilization of AgNPs in the suspension.
Characterization of AgNPs by XRD
The XRD analysis was obtained by a Shimadzu XRD7000 instrument. The AgNPs diameters were determined using Scherrer equation (D= 0.9 λ / β cosθ), were: D = average of particles diameter; λ = wave-length of electromagnetic radiation; θ = diffraction angle; β (2θ) = width at half height of the diffraction peak.
Particle size (DLS) analysis and potential zeta
Particle size and zeta potential (ζ) measurements were carried out in a Malvern Zetasizer Nano, Model ZS 3600. The hydrodynamic diameter was measured by dynamic light scattering with laser of wavelength 633 nm and a fixed scattering angle of 173°. Particle size was measured considering the particle as spherical like. Each sample was measured three times for two replicates samples at a constant temperature of 25 ± 1°C.
Nanoparticle tracking analysis (NTA)
The intensity analyses of nanoparticles in aqueous suspension were visualized using nano sight-nanoparticle tracking analysis (NTA) version 2.0 Release Version Build 0127.
Characterization of AgNPs by transmission electron microscopy (TEM)
The characterization of silver nanoparticles was done using TEM images to determine the size and shape of nanoparticles by a Zeiss Libra 120 instrument at 120 kv.
Antimicrobial activity
Qualitative assay
The agar diffusion assay was used to screen the AgNPs antimicrobial activity against the following microorganisms test: Escherichia coli CBAM 0001, Staphylococcus aureus ATCC 25923, Mycobacterium smegmatis INCQS 061, Candida albicans DPUA 1706, Candida atlantica DPUA 1323 and Candida valderwaltii DPUA 1327. The bacterial cultures were inoculated on Müller Hinton agar (MHA) and the fungi cultures on Sabouraud agar (SAB) using sterilized cotton swabs (6 mm). In each plate, wells were cut out using a sterilized gel borer and 100 μL of biosynthesized AgNPs were used as a test sample against the clinical isolates. Inoculated plates were incubated at 37°C for 24 h. After incubation, the plates were examined for the presence of an inhibition zone around the wells.
Minimum inhibitory concentration (MIC) determination
The antibacterial activity of AgNPs was tested against three bacterial strains: S. aureus ATCC 25923 (Gram-positive), methicillin-resistant S. aureus ATCC 43300 (MRSA) (Gram-positive) and E. coli CBAM 0001 (Gram-negative). Minimum inhibitory concentration (MIC) was determined according to the method of Quelemes et al. (2013) using 96-well micro dilution plates, where the strains (concentration of 5 × 105 CFU/ml) were exposed to two-fold dilution series of AgNPs solution with concentrations ranging from 0.97 to 500 μM. The same procedure was used to determine the MIC of AgNO3 (control). MIC was defined as the lowest concentration of agent that restricted the bacterial visible growth. All assays were performed in triplicate.
Atomic force microscopy analysis (AFM)
The AgNPs extract with the best MIC results was selected for the atomic force microscopy analysis (AFM) using concentrations ranging from 0.975 to 500 µM. A concentrated inoculum of 1 × 108 CFU/mL was used in each well for better observation by AFM. After incubation for 24 h, 30 µL of the culture media containing MIC-treated or MIC-untreated bacteria were deposited into a clean glass surface and dried in bacteriological incubator at 35°C for 10 min. The samples were gently rinsed twice with 1 mL of deionized water to remove salt crystals and dried again at the same described conditions before AFM analysis. All samples were prepared at the same time, exposed to the same conditions and examined within 8 h of deposition. AFM was carried out with a TT-AFM microscope from AFM Workshop (USA). The analysis of the effect of AgNPs on S. aureus cells was carried out in vibrating mode, using NSG10 cantilevers (NT-MDT) with resonant frequency of approximately 280 kHz. Images were analyzed using Gwyddion software 2.33. Multiple areas of each sample were examined and representative images were shown.
Mammalian cell proliferation assay - real-time cell analyzer (RTCA)
To investigate the effects of Streptomyces silver nanoparticles on cell proliferation, murine fibroblast cell line NIH-3T3 (ATCC number 1658) was used. Cells were maintained in 25 cm2 culture flasks (TPP, Switzerland) at 37°C, 5% (v/v) CO2, in DMEM (Dulbecco's Modified Eagle's Medium/Sigma-Aldrich) medium supplemented with 10% (v/v) heat inactivated fetal bovine serum (Invitrogen, USA); 100 IU/mL penicillin and 100 μg/mL streptomycin. The proliferation assays were carried out using Real-Time Cell Analyzer (xCELLigence RTCA DP instrument - Roche). Previously in twenty-four hours, the nanoparticle exposure (t = -24 h) in 5000 cells/well were seeded in E-plates. This first procedure aims to allow cell attachment before treatments. At t=0 h, cells (n=3 well per treatment) were exposed to different silver nanoparticle concentrations (control = 0, 1, 10, 50, 100 µM) and the proliferation pattern was followed for 70 h. The primary results were expressed by a cell index that is correlated with the number of cells attached in plate bottom. It is a strong indication of cell proliferation. The statistical analyses were conducted in two ways:
(1) Comparisons of the area under the curve (AUC) of cell index versus time from the different experimental groups. The analyses of variance (ANOVA) were made followed by Dunnett's test (p < 0.05) among the experimental groups;
(2) Determination of the LC50% (lethal concentration for 50% of exposed cells): for that, a nonlinear curve between the cell index and the log of nanoparticle concentration was calculated for different time points. A curve with some of those results was produced to observe the evolution of LC50% along the exposure time.
Characterization of cultures
The colonies of Streptomyces sp. DPUA 1549, Streptomyces sp. DPUA 1747 and Streptomyces sp. DPUA 1748 were authenticated after reactivation. For Streptomyces sp. DPUA 1549, the mycelium colour varied from white to light grey with pigment production; in Streptomyces sp. DPUA 1747, there was no observed pigment production; and Streptomyces sp. DPUA 1748 showed united colonies without fragmentation, reverse cream, brown mycelium land, without exudate, rough and overlapping edges (Figure 1).
Identification of the microorganisms
The comparison of rDNA sequences is a particularly powerful tool in Streptomycetes family taxonomy (Anderson and Wellington, 2001). Comparison of the 16S rDNA sequences of cultures with the GenBank database showed that these isolates belong to the genus Streptomyces with 100% homology. The sequence of Streptomyces sp. DPUA 1549, Streptomyces sp. DPUA 1747 and Streptomyces sp. DPUA 1748 revealed high similarity with S. parvulus, S. seoulensis and S. owasiensis, respectively.
Synthesis and characterization of nanoparticles
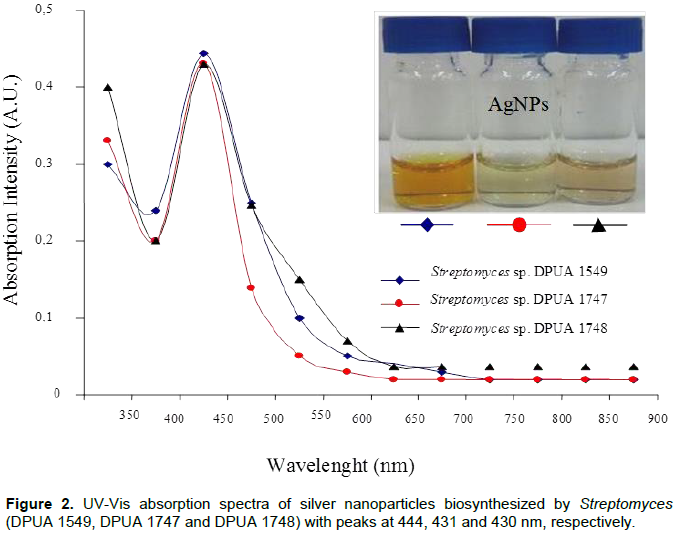
The use of UV visible spectra in the AgNPs characterization is made by observation of absorptions from 420 to 445 nm. These references are attributed to plasmonic bands of silver nanoparticles that present spherical shape (Narasimha et al., 2013) (Figure 2). The visible-UV spectrum of DPUA 1549, DPUA 1747 and DPUA 1748 presented values of lmax close to each other. However, a band enlargement of Streptomyces owasiensis DPUA 1748 was observed, suggesting a dimension rise of the AgNPs obtained. No color change was observed in the culture filtered without silver nitrate. Transmission electronic microscopy has proportioned a detailed view about size and morphology of AgNPs. TEM images of Streptomyces sp. DPUA 1549, DPUA 1747 and DPUA 1748 from aqueous solution are demonstrated in Figure 3A, B and C. The nanoparticles presented spherical shape with larger diameters of DPUA 1748, confirming with data obtained from DLS and UV-vis.
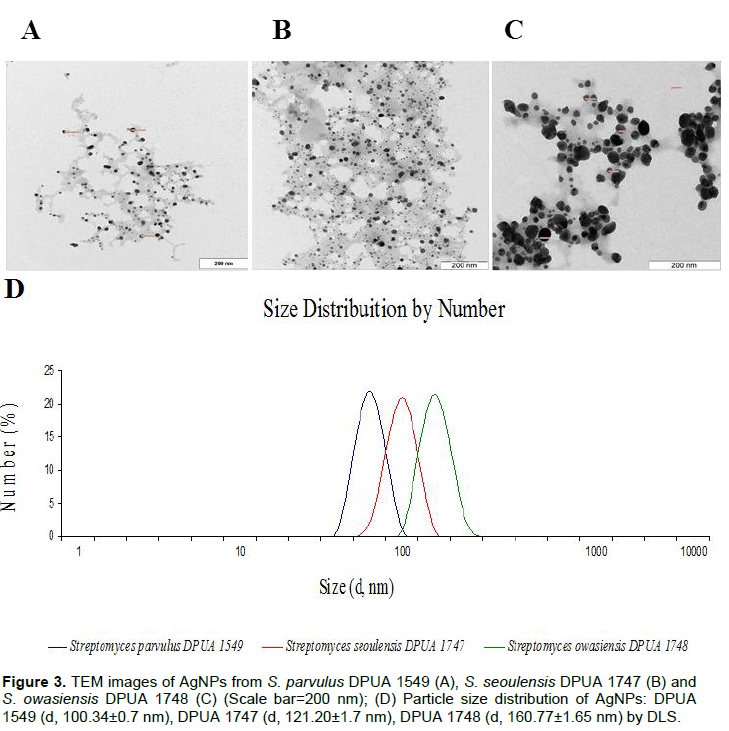
The results obtained in TEM study indicated that the nanoparticles are single spread with average size from 1 to 40 nm. Also, it was observed that the particles involved an organic matrix layer acting as covering agent to the silver nanoparticles (Basavaraja et al., 2008). According to Golinska et al. (2014), nanoparticles stabilization is necessary to avoid aggregation and protect its properties.
Golinska et al. (2014) and Narasimha et al. (2013) reported that metallic nanoparticles biologically synthetized are more stable and disperse better, presenting a secreted coverage by these biological systems. Some studies that used biological systems confirmed the presence of proteins as stabilization agents of metallic nanoparticles (Ingle et al., 2014; Prakasham et al., 2012).
Silver nanoparticles TEM analysis is in agreement with the results presented by Manivasagan et al. (2013), suggesting that the AgNPs were synthetized due to the action of actinomycetes free cells extracts as bio-reducers in this process. Data of DLS demonstrated that the average particles size of Streptomyces sp. DPUA 1549, DPUA 1747 and DPUA 1748 were 100, 121 and 160 nm, respectively. An unimodal distribution and low values of polidispersion to the systems was observed (Figure 3D).
Nanoparticle tracking analysis (NTA) was used to confirm the obtained results by DLS (Figure 4A, B and C) and they are in agreement with all samples analyzed. NTA values were 116, 115 and 120 nm to DPUA 1549, DPUA 1747 and DPUA 1748, respectively, confirming the existence of only one population of nanoparticles without significant size variations between them. The value of zeta potential of silver nanoparticles from Streptomyces sp. DPUA 1549, DPUA 1747 and DPUA 1748 were -20.3, -6.77 and -7.94 mV, respectively (Table 1).
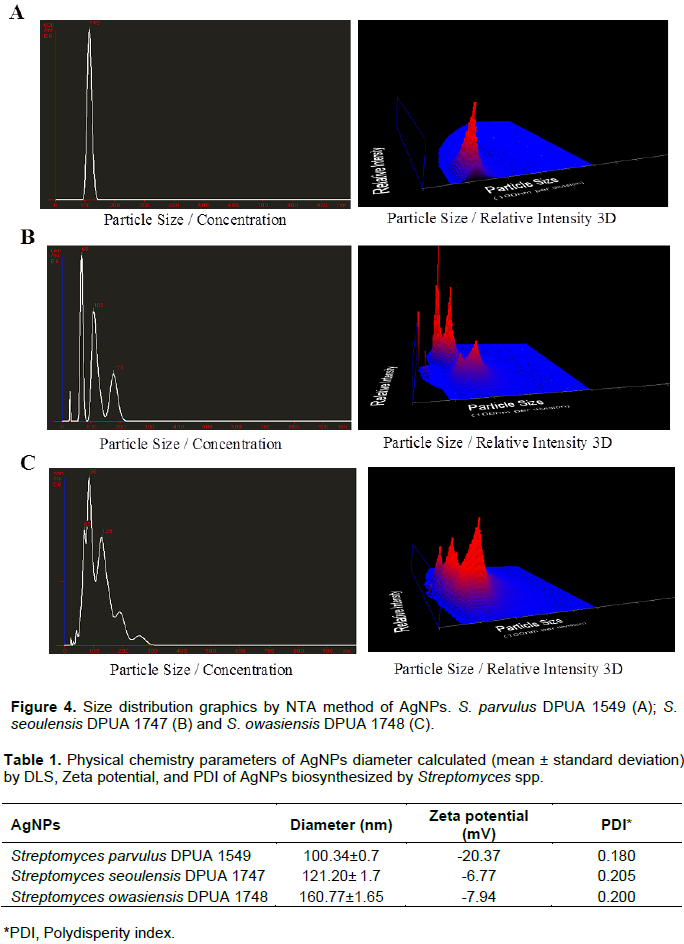
FTIR spectra of DPUA 1549, DPUA 1747 and DPUA 1748 silver nanoparticles presented bands in 3416, 2940, 1634 and 1054 cm-1 that can be attributed to vibrations of symmetric stretching of primary amine, non-symmetric stretching of C-H and stretching due to –C=O and C-O-C groups, respectively. These results also suggest the presence of amino acids and peptides residues which are capable to link to metal, suggesting that biological molecules could play an action in the formation and stabilization of silver nanoparticles in aqueous medium (Basavaraja et al., 2008; Narasimha et al., 2013).
The XRD method was used to determine and confirm the crystalline structure of the synthesized nanoparticles. S. parvulus DPUA 1549 showed peaks in 38°, 44°, 66°, 78° and 81° of 2θ which describes the presence of planes (111), (200), (220), (222) and (311) of face-centred cubic silver nanoparticles structure. The presence of silver oxide in 33° and in 55° was described in the study of Feng et al. (2000). These values are in agreement with those obtained by JCPD (Joint Committee on Powder Diffraction, standard file nº. 04-0783) and a similar XDR pattern with S. seoulensis DPUA 1747. In S. owasiensis DPUA 1748, a low content of silver nanoparticles and silver oxide mixture indicated a low reactivity of this cultured biocompounds with silver nitrate (Table 3).
Antimicrobial activity
Silver (Ag) is a metallic element presenting antimicrobial properties against several microorganisms as fungi, Gram-positive and Gram-negative bacteria. The antimicrobial activity of AgNPs can be more efficient due to their small size particles, high superficial area and volume ratio (Jung et al., 2008; Tamboli and Lee, 2013). The AgNPs from S. parvulus DPUA 1549 and S. owasiensis DPUA 1748 presented antagonist action, in a large spectrum, against all pathogenic microorganisms tested (bacteria and yeasts). The inhibition zones varied from 12 to 34 mm (Table 2) with significant values of DPUA 1549 against S. aureus. The synergic effect of AgNPs promoted an inhibition raise against the microorganisms test. Manivasagan et al. (2013) and Chauhan et al. (2013) proposed that the antibiotic molecules produced by actinomycetes contain several active groups as hidroxile and amide that can easily react with AgNPs by chelation. It suggests that this reaction can raise the growth inhibition of bacterial cells.
MIC assay had the proposal to determine the lowest concentration of AgNPs produced, capable to inhibit microbial growth of pathogens. The AgNPs produced in aqueous solution by S. parvulus DPUA 1549 showed synergic effect with high efficiency in low values of concentration against the tested Gram-negative bacteria E. coli CBAM 001, Gram-positive bacteria S. aureus ATCC 25923 and the methicillin-resistant S. aureus (MRSA ATCC 43300) (Table 3). Besides, an important result is the most effective action of the AgNPs against these Gram-positive bacteria. Other studies demonstrated an antagonistic action against Gram-negative and Gram-positive pathogens (Kumar et al., 2015; Zonooz and Salouti, 2011). The negative zeta potential (-20.37) and size of nanostructures (100 nm) of S. parvulus DPUA 1549 sample was lower when compared to the other tested samples, confirming the results of MIC. According to Kumar et al. (2015) and Kaviya et al. (2011), particles dimensions influence the antimicrobial activity of coloidal AgNPs. Durán et al. (2010) reports that silver nanoparticles obtained in different formulations, sizes and shapes exhibit variable antimicrobial activity.
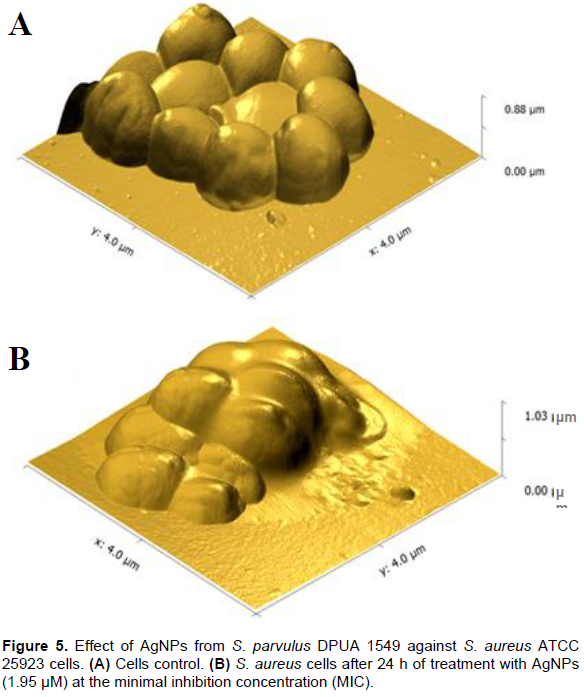
After previous observation of AgNPs antimicrobial action by MIC, the particles of S. parvulus DPUA 1549 were selected to analyze the structural effect against S. aureus ATCC 25923 using atomic force microscopy (AFM). The micrographs of S. aureus ATCC 25923 incubated without AgNPs showed that the bacteria remained structurally intact (Figure 5A) while the bacteria treated with AgNPs lost the structural integrity of the membrane (Figure 5B). Although the antimicrobial action mechanism of AgNPs is not totally understood, many reports demonstrate that silver interaction and bacterial membrane components caused structural damage to these cells (Jung et al., 2008; Pal et al., 2007).
According to research data, there are some possible action mechanisms to the AgNPs activity, and they include: membrane structural damages; decreasing cell viability due interaction with cofactors and ions; inhibition of DNA replication and, consequently, the protein expression; and the inactivation of some essential enzymes to ATP production (Durán et al., 2010). Li et al. (2011) reported that AgNPs act on membrane permeability. After treatment of bacteria with AgNPs, they seemed to suffer lyses occurring from rupture of wall cell resulting in the release of cellular contents to the external environment.
Some studies report the AgNPs action against S. aureus; however, there are no details about all the mechanisms associated to this activity (Jung et al., 2008). For that reason, these particles make it difficult for the microbial ability to enhance defense mechanisms, thus decreasing the resistant strains. Pal et al. (2007) and Shang et al. (2014) affirmed that AgNPs interaction with cell membrane depends on the surface availability. If the nanoparticles are small, the antimicrobial effect can be higher. The AgNPs synthesized by S. parvulus DPUA 1549 might be loaded with antibiotic substances during biosynthesis and produce small bionanoparticles that demonstrate a more efficient antibiotic effect.
Cytotoxic effects - mammalian cell proliferation assay
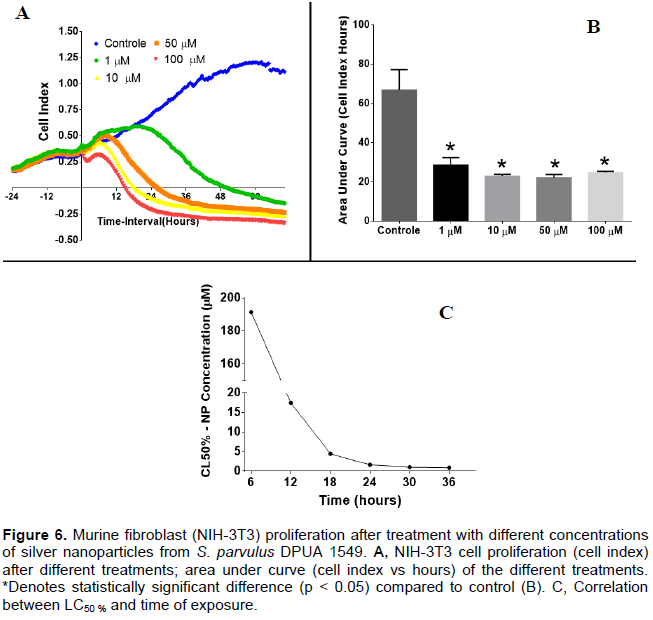
Based on AgNPs antimicrobial activity, cytotoxic activity of the sample was conducted. The mammalian cell proliferation assay after treatment with different concentrations of silver nanoparticles from S. parvulus DPUA 1549 showed that all concentrations evaluated impaired NIH-3T3 cell proliferation. In Figure 6A, it is possible to observe a concentration dependent manner of this proliferation impairment however in the area under curve (AUC) analyses (Figure 6B) no significant differences were observed among the nanoparticle treatments. All of them presented significant reduction (p < 0.05) compared to the control. In addition, time dependent proliferation impairment was observed. In Figure 6C, the reduction of LC50% along the time can be observed. The results indicate that the potential cytotoxic activity of this nanomaterial is also time dependent. Silver nanoparticles used as antimicrobial agent are gaining highlight in applications, especially in medical areas (Vivek et al., 2012).
The AgNPs biosynthesis mediated by Streptomyces expressed antimicrobial activity against bacteria, especially the Gram-positive ones. The silver nanoparticles from S. parvulus DPUA 1549 were the most efficient strain promoting cell superficial damage in tested bacteria; presented low MIC against multiresistant S. aureus and cytotoxic action in the reduction of NIH-3T3 cells proliferation. This scientific evidence demonstrates that the AgNPs synthetized by Streptomyces parvulus DPUA 1549 are effective and can be considered as a promising antimicrobial agent and a cytotoxic agent. This potential favors the application of these nanoparticles in the elaboration of products to be used in medical and cosmetic industries.
The authors have not declared any conflict of interests.
REFERENCES
|
Abd-Elnaby HM, Abo-Elala GM, Abdel-Raouf UM, Hamed MM (2016). Antibacterial and anticancer activity of extracellular synthesized silver nanoparticles from marine Streptomyces rochei MHM13. Egypt. J. Aquat. Res. 42(3):301-312.
Crossref
|
|
|
|
Anderson AS, Wellington EMH (2001). The taxonomy of Streptomyces and related genera. Int. J. Syst. Evol. Microbiol. 51:797-814.
Crossref
|
|
|
|
|
Basavaraja S, Balaji SD, Legashetty A, Rasab AH,Venkatraman (2008). A. Extracellular biosynthesis of silver nanoparticles using the fungus Fusarium semitectum. Mater. Res. Bull. 43:1164-1170.
Crossref
|
|
|
|
|
Bhosale RS, Hajare KY, Mulay B, Mujumdar S, Kothawade M (2015). Biosynthesis, characterization and study of antimicrobial effect of silver nanoparticles by Actinomycetes spp. Int. J. Curr. Microbiol. Appl. Sci. 2:144-151.
|
|
|
|
|
Biglari S, Gholam HS, Gholam RS (2014). Production of gold nanoparticles by Streptomyces djakartensis isolate B-5. Nanomed. J. 1(4):229-237.
|
|
|
|
|
Buszewski B, Railean-Plugaru V, Pomastowski P, Rafi_nska K, Szultka-Mlynska M, Golinska P, Wypij M, Laskowski D, Dahm H (2016). Antimicrobial activity of biosilver nanoparticles produced by a novel Streptacidiphilus durhamensis strain. J. Microbiol. Immunol. Infect. In Press.
Crossref
|
|
|
|
|
Chauhan R, Kumar A, Abraham J (2013). A biological approach to the synthesis of silver nanoparticles with Streptomyces sp JAR1 and its antimicrobial activity. Sci. Pharm. 81:607-621.
Crossref
|
|
|
|
|
Chemat F, Strube J (2015). Green Extraction of Natural Products: Theory and Practice. John Wiley & Sons, Wiley-VCH
|
|
|
|
|
Durán N, Nakazato G, Seabra AB (2016). Antimicrobial activity of biogenic silver nanoparticles, and silver chloride nanoparticles: an overview and comments. Appl. Microbiol. Biotechnol. 100:6555-6570.
Crossref
|
|
|
|
|
Durán N, Marcato PD, De Conti R, Alves OL, Fabio TM, Costa MB (2010). Potential use of silver nanoparticles on pathogenic bacteria, their toxicity and possible mechanisms of action. J. Braz. Chem.
Crossref
|
|
|
|
|
Evelyne RJ, Subbiayh R (2014). Biosynthesis of Silver nanoparticles from Streptomyces olivaceous and its antimicrobial activity. Int. J. Pharma. Res. Health Sci. 2(2):166-172.
|
|
|
|
|
Feng QL, Wu J, Cheng GQ, Cui FZ, Kim TN, Kim JO (2000). A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J. Biomed. Mater. 52(4):662-668.
Crossref
|
|
|
|
|
Golinska P, Wypij M, Ingle AP, Gupta I, Dahm H, Rai M (2014). Biogenic synthesis of metal nanoparticles from actinomycetes: biomedical applications and cytotoxicity. Appl. Microbiol. Biotechnol. 98:8083-8097.
Crossref
|
|
|
|
|
Gul S, Ismail M, Khan MI, Khan SB, Asiri AM, Rahman IU, Khan MA, Kamboh MA (2016). Novel synthesis of silver nanoparticles using melon aqueous extract and evaluation of their feeding deterrent activity against housefly Musca domestica. Asian Pac. J. Trop. Dis. 6(4):311-316.
Crossref
|
|
|
|
|
Ingle AP, Durán N, Rai M (2014). Bioactivity, mechanism of action, and cytotoxicity of copper-based nanoparticles: A review. Appl. Microbiol. Biotechnol. 98:1001-1009.
Crossref
|
|
|
|
|
Jung WK, Koo HC, Kim KW, Shin S, Kim SH, Park Y (2008). Antibacterial activity and mechanism of action of the silver ion in Staphylococcus aureus and Escherichia coli. Appl. Environ. Microbiol. 74(7):2171-2178.
Crossref
|
|
|
|
|
Kaviya S, Santhanalakshmi J, Viswanathan B (2011). Green synthesis of silver nanoparticles using Polyalthia longifolia Leaf extract along with D-Sorbitol. J. Nanotechnol. Volume 2011 (2011), Article ID 152970. 5p.
Crossref
|
|
|
|
|
Kumar PS, Balachandran C, Duraipandiyan V, Ramasamy D, Ignacimuthu S, Al-Dhabi NA (2015). Extracellular biosynthesis of silver nanoparticle using Streptomyces sp. 09 PBT 005 and its antibacterial and cytotoxic properties. Appl. Nanosci. 5:169-180.
Crossref
|
|
|
|
|
Lechevalier MP, Lechevalier HA (1989). Genus Frankia Brunchorst 1886, 174AL, 2410-2417. In. S. T. Williams, M. E. Sharpe, J. G. Holt (Eds.), Bergey's manual of systematic bacteriology, volume 4. The Williams & Wilkins Co., Baltimore.
|
|
|
|
|
Li W, Xie X, Shi Q, Duan S, Ouyang Y, Chen Y (2011). Antibacterial effect of silver nanoparticles on Staphylococcus aureus. Biometals 24:135-141.
Crossref
|
|
|
|
|
Manikprabhu D, Lingappa K (2013). Antibacterial activity of silver nanoparticles against methicillin-resistant Staphylococcus aureus synthesized using model Streptomyces sp. pigment by photo-irradiation method. J. Pharm. Res. 6(2):255-260.
Crossref
|
|
|
|
|
Manivasagan P, Venkatesan J, Senthilkumar K, Sivakumar K, Kim S (2013). Biosynthesis, antimicrobial and cytotoxic effect of silver nanoparticles using a novel Nocardiopsis sp. MBRC-1. BioMed. Research International. Article ID 287638. 9p.
|
|
|
|
|
Narasimha G, Janardhan AM, Khadri H, Mallikarjuna K (2013). Extracellular synthesis, characterization and antibacterial activity of silver nanoparticles by actinomycetes isolative. Int. J. Nano Dimens. 4:77-83.
|
|
|
|
|
Pal S, Tak YK, Song JM (2007). Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the Gram-negative bacterium Escherichia coli. Appl. Environ. Microbiol. 73:1712-1720.
Crossref
|
|
|
|
|
Pantidos N, Horsfall LE (2014). Biological synthesis of metallic nanoparticles by bacteria, fungi and plants. J. Nanomed Nanotechnol. 5(5):1.
Crossref
|
|
|
|
|
Prakasham RS, Buddana SK, Yannam SK, Guntuko SK (2012). Characterization of silver nanoparticles synthesized by using marine isolate Streptomyces albidoflavus. J. Microbiol. Biotechnol. 22:614-621.
Crossref
|
|
|
|
|
Queiroz GM, Silva LM, Pietro LCR, Salgado HRN (2012). Multirresistência microbiana e opções terapêuticas disponíveis. Rev. Bras. Clin. Med. São Paulo 10(2):132-138.
|
|
|
|
|
Quelemes PV, Araruna FB, De Faria BEF, Kuckelhaus SAS, Da Silva DA, Mendonça RZ, Eiras C, Soares MJS, Leite JRSA (2013). Desenvolvimento e atividade antibacteriana de nanopartículas de prata à base de goma do cajueiro. Int. J. Mol. Sci. 14:4969-4981.
Crossref
|
|
|
|
|
Roco MC, Mirkin CA, Hersam MC (2011). Nanotechnology research directions for societal needs in 2020: summary of international study. J. Nanopart. Res. 13(3):897-919.
Crossref
|
|
|
|
|
Samundeeswari A, Dhas SP, Nirmala J, John SP, Mukherjee A, Chandrasekaran N (2012). Biosynthesis of silver nanoparticles using actinobacterium Streptomyces albogriseolus and its antibacterial activity. Biotechnol. Appl. Biochem. 59:503-507.
Crossref
|
|
|
|
|
Schneid AC, Roesch EW, Sperb F, Matte U, da Silveira NP, Costa TMH, Benvenutti EV, de Menezes EW (2014). Silver nanoparticle–ionic silsesquioxane: a new system proposed as an antibacterial agent. J. Mater. Chem. B, 2, 1079.
Crossref
|
|
|
|
|
Selvakumar P, Prakash S, Jasminebeaula S, Uloganathan R (2012). Antimicrobial activity of extracellularly synthesized silver nanoparticles from marine derived Streptomyces rochei. Int. J. Pharm. Biol. Sci. 3:188-197.
|
|
|
|
|
Shang L, Nienhaus K, Gerd UN (2014). Engineered nanoparticles interacting with cells: size matters. J. Nanobiotechnol. 12(5):2-11.
Crossref
|
|
|
|
|
Silva-Vinhote NM, Marinho-Pereira T, Astolfi-Filho S, Matsuura T (2011). Taxonomic characterization and antimicrobial activity of actinomycetes associated with foliose lichens from the amazonian ecosystems. Austral. J. Basic Appl. Sci. 5:910-918.
|
|
|
|
|
Tamboli DP, Lee DS (2013). Mechanistic antimicrobial approach of extracellularly synthesized silver nanoparticles against gram positive and gram negative bacteria. J. Hazard. Mater. 260:878-884.
Crossref
|
|
|
|
|
Vivek R, Thangam R, Muthuchelian K, Gunasekaran P, Kaveri K, Kannan S (2012). Green biosynthesis of silver nanoparticles from Annona squamosa leaf extract and its in vitro cytotoxic effect on MCF-7 cells. Process Biochem. 47:2405-2410.
Crossref
|
|
|
|
|
Waksman SA, Woodruff HB (1941). Actinomyces antibioticus a new soil organism antagonistic to pathogenic and non-pathogenic bacteria. J. Bacteriol. 42:231-249.
|
|
|
|
|
Zonooz NF, Salouti M (2011). Extracellular biosynthesis of silver nanoparticles using cell filtrate of Streptomyces sp. ERI-3. Sci. Iran 18(6):1631-1635.
Crossref
|
|