ABSTRACT
Contamination of soil with heavy metals is one of the major environmental problems in many countries that reach from many sources as power stations, application of metal pesticides, fertilizers, and sewage sludge. In this study, ten Rhizobium leguminosarum bv. viciae isolates were collected from different governorates in Egypt (Menoufia, Kafr El-Sheikh, Qalubia, Fayoum, Ismailia, Sharkya, Dakhalia, Behira and North Sinai) to evaluate their resistance to three heavy metals (Cu, Zn and Pb). The results showed that the isolates of RL3 and RL6 exhibited the best resistance toward the heavy metals tested. PCR based specific primers were used to screen the tested isolates for detection of some heavy metal resistant genes (copA, pbrA and czcD). The Pb-resistant gene pbrA was detected in most of tested isolates except RL7 and RL9 isolates, while, the Cu-resistant gene copA was found in all isolates except RL1, RL2, RL4 and RL8 isolates; however, the Zn-resistant gene czcD was detected only in the RL9 isolate. SDS-PAGE analysis was used to study the protein banding patterns for some tested isolates under lead stress and compared them with their untreated control.
Key words: CopA, pbrA and czcD genes, heavy metals, Rhizobium.
Rhizobia are Gram-negative soil bacteria with high agronomic significance due to their ability to establish nitrogen-fixing symbiosis with leguminous plants through invading their roots and forming nodules for atmospheric nitrogen fixation (Stan et al., 2011). The symbiosis process can be affected by many environmental factors such as temperature, soil acidity and salinity (Dart, 1977; Gibson and Jordan, 1983; Sobti et al., 2015). Heavy metals soil contamination is among the factors that have negative effects on the growth of each rhizobium and plant. Nowadays, with increasing industrial activities, the use of industrial waste waters for irrigation and application of metal containing pesticides and fertilizers, the level of soil pollution with heavy metals is increased (Gopalakrishnan et al., 2014; Stan et al., 2011). Approximately 30% of the terrene environment is suggested to be degraded or contaminated and this surely can cause disaster problems for each environment and agricultural production (Alloway and Trevors, 2013; Valentín et al., 2013). The exposure to heavy metals is toxic not only for soil microorganisms but also for plants. It has been showed that with increasing concentrations of heavy metals such as Cu, Zn and Pb, the bacterial counts of Rhizobium sp. are reduced and also the expression of nod genes was varied (Stan et al,. 2011; Chaudri et al., 2008). On the contrast, it was suggested that Rhizobia can tolerate high heavy metal concentrations in different ways and may play a significant role in the restoration of contaminated soil (Carrasco et al., 2005; Teng et al., 2015). The symbiotic relationship between rhizobia and legumes reinforce elimination rate of pollutants (Glick, 2010). Hao et al. (2014) showed that rhizobia heavy metal tolerance mechanisms may include: (i) Adsorption and accumulation of heavy metals; and (ii) microbial secretion of enzymes and bioactive metabolites to increase their bioavailability and sequester their toxicity.
In this study, ten rhizobial isolates were collected from root nodules of broad bean (Vicia faba L.) plants representing different geographic sites in Egypt. The objectives of this study was to: (i) characterize these isolates by comparing their growth on medium supplemented with different concentrations of heavy metals (Cu, Pb and Zn); and (ii) screen the tested isolates for presence of heavy metal resistance genes using polymerase chain reaction (PCR) and to study the protein banding patterns under some heavy metal stress using sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE).
Isolation of Rhizobia
Ten isolates of Rhizobium leguminosarum bv. Viciae (RLV) were collected from root nodules of V. faba L. plants representing several Egyptian governorates according to the methods described by Vincent, 1970. Table 1 shows the isolation sites and the name of the isolates.
DNA extraction
Total genomic DNA was extracted from bacterial cultures grown in yeast extract mannitol media (YEM) as described by Shamseldin et al. (2009). The quality and quantity of DNA was characterized both spectrophotometrically and by 0.8% agarose gel. The DNA from all isolates produced clear sharp bands, indicating good quality of DNA.
Amplification of heavy metals resistant genes
The primers for the amplification of a Cu-resistance gene (pcoR) were pcoRf; 5-CAGGTCGTTACCTGCAGCAG-3(forward) and pcoRr; 5- CTCTGATCTCCAGGACATATC -3(reverse) (Trajanovska et al., 1997). Pb-resistant gene (pbrA) primers were pbrA1; 5-ATG AGCGAATGTGGC TCGAAG-3(forward) and pbrA2; 5-TCATCGACGC AACAGCCTCAA-3(reverse) (Borremans et al., 2001). Zn-resistant gene (czcD) primers were czcD1; 5-CAGGTC ACTGAC ACG ACC AT-3(forward) and czcD1; 5-CAT GCT GAT GAG ATT GAT GAT C-3(reverse) (Nies et al., 1989). PCRs were carried out in 25 µl reaction mixtures using the following conditions: Initial denaturation at 94°C for 10 min, followed by 35 cycles of 94°C for 1 min, 60°C for 1 min, and 3 min at 72°C.A final extension was done at 72°C for 7 min. PCR products were separated on 2% agarose gels at 100 V for 1 h in TBE buffer, stained with ethidium bromide, and photographed under UV light.
Evaluation of heavy metals tolerance
The RLV isolates were evaluated for their tolerance against three different heavy metals (Cu, Zn and Pb) by plating in YEMA medium. The stock solutions of heavy metals (mM) were added to sterile agar as follows: CuCl2.2H2O 0.5, 1 and 2; ZnSO4.7H2O0.5, 1 and 2; Pb(C2H3O2)2.3H2O 0.5, 1.0 and 2.The plates were inoculated with bacterial cells and the bacterial growth was evaluated after 7 days at 28°C (Ausili et al., 2002). Isolates were considered resistant if growth was observed or sensitive if otherwise.
Protein banding patterns of Rhizobial isolates
The cultures of tested isolates growing on Broth YEM medium and supplemented with 0.5 mM of Pb(C2H3O2)2.3H2O were pelleted and Sodium dodecyl sulfate- polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described by Pereira et al. (2006). Gels were stained with Coomassie brilliant blue R-250 and photographed.
Detection of some heavy metal resistant genes
In this study, PCR based on heavy metal specific primers was used to screen and detect some heavy metal resistant genes in the tested isolates (copA, pbrA and czcD). The primers used for the amplification Pb-resistant gene pbrA yielded a band of approximately 500 bp in most of tested isolates except RL7 and RL9 (Figure 1A). For Cu-resistant gene copA, the PCR produced one band of approximately 650 bpin most of tested isolates except RL1, RL2, RL4 and RL8 (Figure 1B). This band is homologous to that obtained by Trajanovska et al. (1997).
On the other side, only one band of approximately 500 bp was detected in RL9 isolate with amplification of the Zn -resistant gene czcD (Figure 1C). It must be mentioned that microbes use heavy metal resistant genes to encode products that reduce or eliminate the toxicity of heavy metals for adaptation of the different environment stresses (Wei et al., 2009). From these genes, pbrA, which encodes a P-type Pb(II) efflux ATPase in the lead resistance operon that involved in uptake, efflux, and accumulation of Pb(II) (Borremans et al., 2001). In addition, CopA is essential gene in copper resistance operon and catalyzes the intake of copper (Wei et al., 2009). Moreover, the CzcDis necessary gene in Zn-resistant operon and mutation in this gene can disrupt level of Zn-resistance (Nies et al., 1989).
Screening of Rhizobium isolates for heavy metals resistance
All of Rhizobium isolates were tested for their resistance to heavy metals using concentrations of 0.5, 1 and 2 mM of Cu, Pb and Zn (Table 2). The isolates were considered to be resistant when the growth occurs in the presence of heavy metals or sensitive if otherwise. First with Pb treatment, all isolates were able to grow at the low concentration (0.5 mM), while most of the isolates were found to be resistant to Pb at 1 mM except RL2, RL8 and and RL9 isolates. At the highest concentration of lead (2 mM), most of isolates failed to grow except RL3 and RL6 isolates. For Cu treatment, all isolates were able to grow at the low concentration (0.5 mM) except RL8 and RL10 isolates. Moreover, only the isolates RL1, RL4 and RL6 were succeeded to grow at concentration of 1 mM of Cu, while no isolates appeared at the highest concentration of Cu (2 mM). Finally, for Zn treatment, all of isolates succeeded to grow at 0.5 mM except the isolate RL7, while most of isolates can grow at 1 mM of Zn except RL7, RL8 and RL10 isolates. Furthermore, no isolates appeared at the highest concentration of Zn (2 mM) in general, the isolates RL6 and RL 3 showed the highest levels of Pb resistance, while RL1, RL 4 and RL6 isolates exhibited the highest levels of Cu resistance. In addition, RL3, RL 5, RL6 and RL9 were the best isolates in Zn resistance. It must be mentioned that the ability to resist the heavy metals decreased with increasing their concentrations. These results are consistent with previous studies shown that the increased concentrations of heavy metals can affect the growth, morphology and activities of microorganisms in nitrogen fixation (Khan and Scullion, 2002; Lakzian et al., 2002; Shi et al., 2002; Pereira et al., 2006). Furthermore, the tested isolates exhibited more sensitive against Cu than the other heavy metals Pb and Zn. These observations are in agreement with previous studies showed that the high concentrations of Cu are toxic to soil microorganisms by affecting their structural diversity and metal tolerance (Cervantes and Gutierrez-Corona, 1994; Dell’Amico et al., 2008).
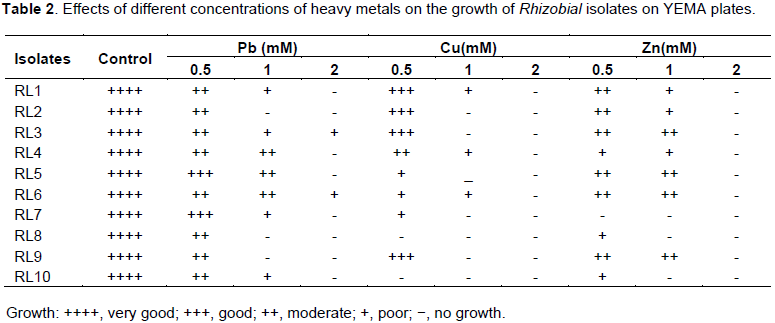
The appearance of resistance levels in isolates as RL 3 and R6 against some heavy metals as Pb is due to these isolates contain the pb-resistant gene pbrA (Figure 1A). On the other hand, the appearance of resistance levels in some isolates although their resistance genes did not detected by PCR is not understood. One of explanations is that these isolates were isolated from soil polluted with heavy metals and have probably adapted this environmental stress. This explanation is supported with previous literature shown that the selective pressure of metals on microorganisms can lead to microbial populations with a high resistance to metals (Pereira et al., 2006). Another explanation, these isolates may have other resistance mechanisms for removal of these elements (Wei et al., 2009; Teng et al., 2015).
Characterization of Rhizobium isolates by SDS-PAGE
The analysis of protein alterations seems to be a good indicator to estimate the level of stress imposed to Rhizobium populations (Pereira et al., 2006). Hence, SDS-PAGE analysis was used to study the protein banding patterns for some Rhizobium isolates under lead stress and compared them with their untreated control. The isolates RL2 and RL8 were selected as sensitive isolates, while the isolates RL3 and RL 6 were selected as resistant according to their growth under lead stress (Figure 2). In general, Rhizobium isolates showed similar banding patterns however some differences were detected as indicated in Figure 2. Comparative analysis of the lanes showed the absence of two protein bands (about 25 and 40 kDa) in the sensitive isolate RL2 (treated and their control) compared to other isolates.
Isolation of rhizobia strains resistant to stresses like heavy metals is very important for efficient nitrogen fixation and improving plant productivity especially in the contaminated areas. The results of this study showed in general that the isolates of RL3 and RL6 found to be the best isolates to tolerate Pb, Cu and Zn heavy metal elements. Future studies must be done to test these isolates in fields contaminated with heavy metals for increasing nitrogen fixation level by faba bean plants cultivated in contaminated soils.
The authors have not declared any conflict of interests.
REFERENCES
|
Alloway BJ, Trevors JT (2013). Heavy Metals in Soils-Trace Metals and Metalloids in Soils and their Bioavailability. New York: Springer Dordrecht Heidelberg.
|
|
|
|
Ausili P, Borisov A, Lindblad P, Martensson A (2002). Cadmium affect the interaction between peas and root nodule bacteria. Acta Agric. Scand. B Soil Plant Sci. 52:8-17.
|
|
|
|
|
Borremans B, Hobman JL, Provoost A, Brown NL, van der Lelie D (2001).Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. J. Bacteriol.19:5651-5658.
Crossref
|
|
|
|
|
Carrasco JA, Armario P, Pajuelo E, Burgos A (2005). Isolation and characterisation of symbiotically effective Rhizobium resistant to arsenic and heavy metals after the toxic spill at the Aznalcollar pyrite mine. Soil Biol. Biochem. 37:1131-1140.
Crossref
|
|
|
|
|
Cervantes C, Gutierrez-Corona F (1994). Copper resistance mechanisms in bacteria and fungi. FEMS. Microbiol. Rev. 14:121-138.
Crossref
|
|
|
|
|
Chaudri A, McGrath S, Gibbs P, Chambers B, Carlton-Smith C, Bacon J, Campbell C, Aitken M (2008). Population size of indigenous Rhizobium leguminosarum biovar trifoliiin long-term field experiments with sewage sludge cake, metal amended liquid sludge or metal salts: Effects of zinc, copper and cadmium. Soil Biol. Biochem. 40:1670-1680.
Crossref
|
|
|
|
|
Dart P (1977). Infection and development of leguminous nodules. In. A Treatise on Di nitrogen Fixation, Section III: Biology ed. Hardy.
|
|
|
|
|
Dell'Amico E, Mazzocchi M, Cavalca L, Allievi L,Andreoni V (2008). Assessment of bacterial community structure in a long-term copper-polluted exvineyard soil. Microbiol. Res.163:671-683.
Crossref
|
|
|
|
|
Gibson AH, Jordan DC (1983). Eco physiology of nitrogen fixing systems.In Physiological Plant Ecology III. Responses to the Chemical and Biological Environment ed. Lange, O.L., Nobel, P.S., Osmond, C.B. and Zeigler, H. Berlin: Springer-Verlag. Pp. 301-390.
|
|
|
|
|
Glick BR (2010). Using soil bacteria to facilitate phytoremediation. Biotechnol. Adv. 28:367-374.
Crossref
|
|
|
|
|
Gopalakrishnan S, Sathya A, Vijayabharathi R, Varshney RK, Gowda CLL, Krishnamurthy L (2014). Plant growth promoting rhizobia: challenges and opportunities. 3 Biotech 5(4):355-377.
|
|
|
|
|
Hao X, Taghavi S, Xie P, Orbach MJ, Alwathnani HA, Rensing C, Wei G (2014). Phytoremediation of heavy and transition metals aided by legume-rhizobia symbiosis. Int. J. Phytoremediat. 16:179-202.
Crossref
|
|
|
|
|
Khan M, Scullion J (2002). Effects of metal (Cd, Cu, Ni, Pb or Zn) enrichment of sewage-sludge on soil microorganisms and their activities. Appl. Soil Ecol. 20:145-155.
Crossref
|
|
|
|
|
Lakzian A, Murphy P, Turner A, Beynon JL, Giller KE (2002). Rhizobium leguminosarumbv. viciaepopulations in soils with increasing heavy metal contamination: abundance, plasmid profiles, diversity and metal tolerance. Soil Biol. Biochem. 34:519-529.
Crossref
|
|
|
|
|
Nies DH, Nies A, Chu L, Silver S (1989). Expression and nucleotide sequence of aplasmid determined divalent cation effux system from Alcaligeneseutrophus. Proc. Natl. Acad. Sci. 86:7351-7355.
Crossref
|
|
|
|
|
Pereira SIA, Lima AIG, Figueira EMDAP (2006). Heavy metal toxicity in Rhizobium leguminosarum biovar viciae isolated from soils subjected to different sources of heavy-metal contamination: effects on protein expression. Appl. Soil. Ecol. 33:286-293.
Crossref
|
|
|
|
|
Shamseldin A, El-saadani M, Sadowsky MJ, An CS (2009). Rapid identification and discrimination among Egyptian genotypes of Rhizobium leguminosarum bv. viciae and Sinorhizobium meliloti nodulating faba bean (Vicia faba L.) by analysis of nodC, ARDRA, and rDNA sequence analysis. Soil Biol. Biochem. 41:45-53.
Crossref
|
|
|
|
|
Shi W, Bischoff M, Turco R, Konopka A (2002).Long-term effects of chromium and lead upon the activity of soil microbial communities. Appl. Soil Ecol. 21:169-177.
Crossref
|
|
|
|
|
Sobti S, Belhadj HA, Djaghoubi A (2015). Isolation and Characterization of The Native RhizobiaUnder Hyper-Salt Edaphic Conditions in Ouargla (southeast Algeria); international Conference on Technologies and Materials for Renewable Energy, Environment and Sustainability, TMREES 15.Energy Procedia. 74:1434-1439.
Crossref
|
|
|
|
|
Stan V, Gament E, Cornea CP, Voaides C, Dusa M, Plopeanu G (2011). Effects of heavy metal from polluted soils on the Rhizobium diversity. Not. Bot. Horti. Agrobot. Cluj. Napoca 39:88-95.
|
|
|
|
|
Teng Y, Wang X, Li L, Li Z, Luo Y (2015). Rhizobia and their bio-partners as novel drivers for functional remediation in contaminated soils. Front. Plant Sci. 6:32.
Crossref
|
|
|
|
|
Trajanovska S, Britz ML, BhaveM (1997). Detection of heavy metal ion resistance genes in Gram-positive and Gram-negative bacteria isolated from a lead-contaminated site. Biodegradation 8:113-124.
Crossref
|
|
|
|
|
Valentín L, Nousiainen A, Mikkonen A (2013). Introduction to organic contaminants in soil: concepts and risks, In. Emerging Organic Contaminants in Sludges: The Handbook of Environmental Chemistry, eds A.G. Kostianoy and D. Barceló (Berlin, Heidelberg: Springer-Verlag). pp. 1-29.
Crossref
|
|
|
|
|
Vincent JM (1970). A manual for the practical study of root nodule bacteria. IBP Handbook No.15, Blackwell Scientific Publications.
|
|
|
|
|
Wei G, Fan L, Zhu W, Fu Y, Yu J, Tang M (2009).Isolation and characterization of the heavy metal resistant bacteria CCNWRS33-2isolated from root nodule of Lespedeza cuneatain gold mine tailings in China. J. Hazard. Mater.162:50-56.
Crossref
|
|