ABSTRACT
Rosa damascena Mill. as a main economic crop in the world is planted for beauty and essential oil production in Ta’if region. For the management and improvement of this important crop, genetic variability was evaluated amongst six Rosa genotypes grown in different plantations using microsatellite (simple sequence repeats, SSR), inter simple sequence repeat (ISSR) and biochemical markers. The six SSR primers showed low level of variation, whereas all ISSR primers generated high levels of polymor-phism ranging from 66.7 to 100%. The biochemical mar¬kers revealed slight polymorphism between the three Rosa species under study. The dendrogram resulted from the combined data of SSR and ISSR splits the 6 genotypes into two main clusters. The first comprised the four R. damascena accessions, and the second grouped R. damascene, Trigintipetala‘and R. hybrid together. ISSR markers can be recommended for the genetic variability analysis in Rosa genome.
Key words: Rosa damascena, microsatellite, inter simple sequence repeat, dendrogram, genetic relationship.
In Saudi Arabia, Rosa damascena is mainly grown for the production of oil that could be considered one of the most expensive oils in the world (Farooq et al., 2013). Despite the history of R. damascena, some doubt has been cast upon the source and origin of this species. A lot of roses have been selected and recombined to produce R. damascena in the Middle East regions including Ta’if (Widrlechner, 1981). Hence, there is consistent need for evaluating the genetic differences among Rosa species and cultivars to provide a continuous development of new germplasm for rose breeding programs and rose oil industry in Ta'if.
Molecular markers could aid breeding by providing dependable tools to investigate the variability among parents and their progenies (Rusanov et al., 2005). Although, Rosa plants have high levels of polyphenols and polysaccharides that make extraction of protein and DNA difficult (Kaul et al., 2009), significant efforts have been done to use these markers for identification, gene expression and biodiversity in Rosa lines, cultivars and hybrids, such as proteins (Dafny-Yelin et al., 2005), isozymes (Grossi et al., 1997; Jayasree et al., 1998), RAPDs (Kiani et al., 2008; Mirzaei and Rahmani, 2011), inter simple sequence repeat (ISSR) (Jabbarzadeh et al., 2010) and simple sequence repeats (SSR) (Stafne et al., 2005; Babaei et al., 2007; Farooq et al., 2013; Nadeem et al., 2014).
Among molecular markers, microsatellite or simple sequence repeats and ISSR as rapid techniques are useful in several areas of plant genetic studies. ISSR-PCR produces variable patterns of loci that are reproducible, abundant and polymorphic (Bornet and Branchard, 2004). Furthermore, SSR markers as codominant factors are able to reveal many alleles that can discriminate among closely related hybrids and cultivars (Nadeem et al., 2014). However, SSRs may be expensive to produce and laborious in particular species. So SSR primers developed from one species could be used for other studies on related species and genera (Stafne et al., 2005).
To increase our understanding of the genetic relationships between Rosa species, we used SSR, ISSR and biochemical markers in genetic diversity analysis of four R. damascena accessions and two related species; R. damascena ØŒTrigintipetala‘ and R. hybrida grown in Ta'if region, Saudi Arabia.
Plant materials
Roses were obtained from different sites of Ta'if region in Saudi Arabia (Table 1). In this study, six genotypes (four accessions of R. damascene and two related species; R. damascena ‘Trigintipetala’, R. hybrida) were investigated.
DNA extraction
Young leaves (0.5 g) of genotypes were taken for DNA extraction. Rose DNA extraction was done based on the cetyl trymethyl ammonium bromide (CTAB) method as described by Doyle and Doyle (1987).
SSR and ISSR primers
In this study, SSR and ISSR primers were selected from previous studies of Zhang et al. (2006) and Jabbarzadeh et al. (2010) (Tables 2 and 3). Fourteen primers were selected that produced good banding patterns for analysis.
PCR conditions
Amplifications were done with a 13 µL total reaction/sample that included 10 µL Taq Master Mix, 1 µL each, forward and reverse primers, and 1 µL DNA. Thermal cycling was done on a Techne TC-3000 (Barloworld Scientific, Ltd. Staffordshire, UK) with the following program: 105°C heated lid, initial denaturation of 94°C for 5 min, and 35 cycles of 94°C for 1 min, 58°C (changed according to primer) for 1 min, 72°C for 1 min, finishing with a final extension of 72°C for 5 min and a final hold at 4°C. Products were confirmed on an agarose gel and stained by ethidium bromide then observed under UV light and photographed by gel documentation system.
Isozyme analysis
The isozymes; α- and β-esteras (EST), acid phosphatase (ACP), alcohol dehydrogenase (ADH) and peroxidase (PRX) were separated in 10% native-polyacrylamide gel electrophoresis as described by Stegemann et al. (1987). For gels staining, protocols of Scandalios (1964) were used for α- and β-EST, Wendel and Weeden (1989) for ACP, Weeden and Wendel (1990) for ADH and Heldt (1997) for PRX. Gels were washed two or three times with tap water; fixed in ethanol: 20% glacial acetic acid and photographed.
Protein analysis
To extract proteins, 1 g of leaves of each genotype was mixed with 1 M Tris-HCl buffer, pH 8.8 and homogenized using a mortar and pestle. After centrifugation, the supernatants were transferred to new tubes and kept in deep-freeze until use. Electrophoresis was carried out by the modified discontinuous sodium dodecyl sulphate-polyacrylamide gel electrophoresis (DISC SDS-PAGE) method (Laemmli, 1970). Gels were stained overnight in coomassie brilliant blue-R250 solution then destained and photographed.
Data analysis
The bands of SSR, ISSR and protein were evaluated by comparing with 100 bp DNA ladder and blue wide range prestained protein ladder (Cleaver Scientific Ltd, UK), respectively, using gel analyzer program (version 3). To determine the genetic relationship among Rosa genotypes, bands of SSR and ISSR patterns were treated as a unit character and coded 1 or 0 for their presence or absence, respectively. Clustering was performed using UPGMA procedure and represented in a phenogram by using SAHN and TREE modules, respectively. The NTSYS-pc 2.2 (Numerical Taxonomy and Multivariate Analysis System, Exeter Software) program was utilized in all previous analysis (Rohlf, 1998).
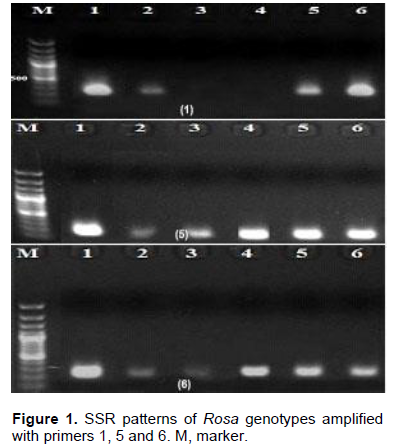
For management and successful improvement of the rose crop, genetic variation within different accessions, cultivars and species of Rosa is needed (Kaul et al., 2009). Of the nine specific SSR primers tested in Rosa, six produced clear and valid products (Table 2). The six primers produced six bands with molecular sizes that ranged from 150 to 422 bp as shown in Figure 1. Additionally, this study has shown that the annealing temperatures for the 6 SSR primers varied from 52-58°C (Table 2). Notably, only 2 out of 6 primers showed low level of variation among rose genotypes tested. These findings contrast that of Zhang et al. (2006) who observed that SSRs revealed a significant level of variation and could differentiate easily among rose cultivars studied. On the other hand, they are in accordance with previous studies depending on SSR markers that did not exhibit any variability among cultivars of R. damascena in Turkey and Bulgaria (Baydar et al., 2004; Rusanov et al., 2005). Although, SSR markers give rapid data from small amount of plant sample, they are costly to produce and can be very exhausted in particular species (Stafne et al., 2005).
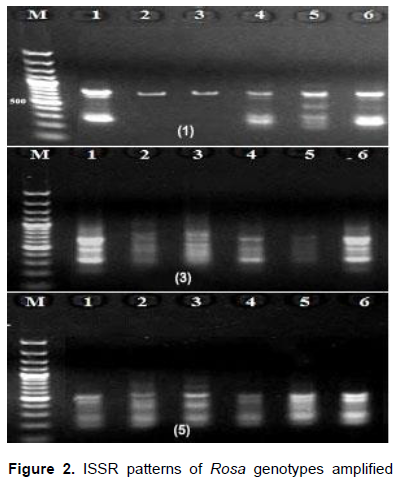
For further evaluation of the genetic relatedness among Rosa genotypes, eight ISSR primers ranging between 11 and 20 bases were used. The annealing temperatures for the eight ISSR primers varied from 36-53°C (Table 3). A total of 37 bands were amplified from the genotypes, of which 34 were polymorphic. The bands sizes ranged from 235-1100 bp (Figure 2). The ISSR patterns revealed 11 unique bands; 2 in R. damascene, 3 in R. damascena ØŒTrigintipetala‘, six in R. hybrid. All primers generated high levels of polymorphism ranging from 66.7 to 100%. These results were in conformity with those of the Pakistani and Iranian scientists that scored high levels of diversity among rose genotypes collected from different regions from Pakistan and Iran (Mirali et al., 2012).
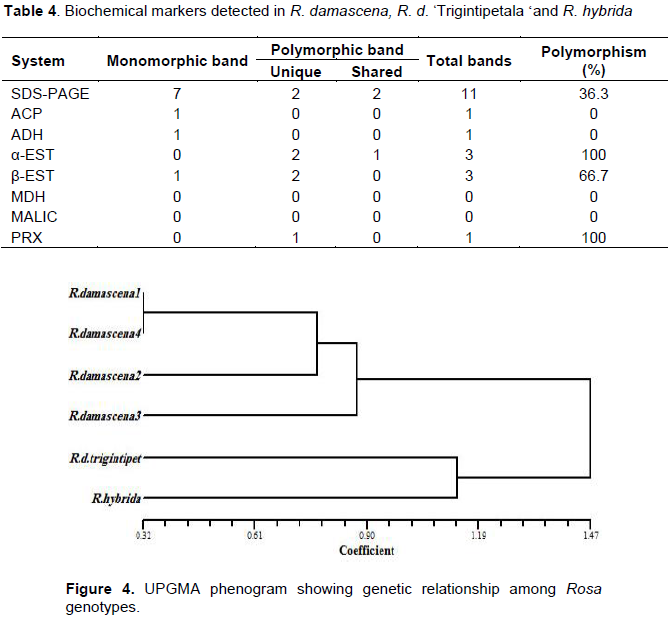
The data of SSR and ISSR were combined to estimate the genetic similarity values and generate a dendrogram showing the relationship among rose genotypes under study. The similarity matrix showed that the highest value (0.966) was between two accessions of R. damascena (collected from Ta'if city and Misan), whereas the lowest value (0.333) was found between one accession of R. damascena and R. hybrida (data not shown). The resultant dendrogram grouped the six genotypes into two main clusters. The first comprised the four R. damascena accessions, and the second grouped R. damascena ØŒTrigintipetala‘and R. hybrid together (Figure 3). Occurrence of genetic variations among accessions of R. damascena might be due to the geographical differences that might cause an evolution through genetic drift, mutation and recombinations. The climatic conditions can also affect and lead to variability within and between Rosa species (Farooq et al., 2013).
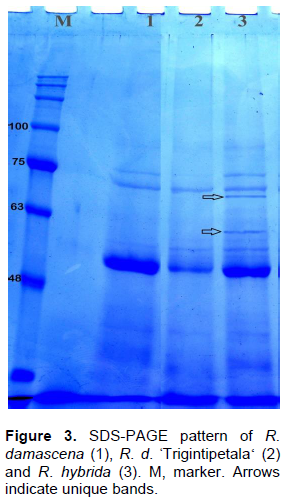
Two biochemical approaches; isozyme and SDS-PAGE, were used to discriminate among R. damascena, R. damascena ØŒTrigintipetala‘and R. hybrida. Seven enzyme systems were used as mentioned in Table 4. The three species did not generate any bands for the two MDH and malic isozymes. One to three bands per isozyme were produced for the remaining five systems. R. damascena ØŒTrigintipetala‘ was distinguished by three unique bands for estesrases, whereas R. damascena and R. hybrida were severally characterized by only one unique band for PRX and β-EST systems, respectively. On the other hand, the produced SDS-PAGE of leaf protein profile of R. damascena, R. damascena ØŒTrigintipetala‘and R. hybrid is shown in Figure 4. A total number of 11 bands were recorded. Molecular weight (Mw) of the protein subunits ranged from 34.7 to 91.5 kDa. The profile revealed seven monomorphic bands in all species and four polymorphic bands including two unique bands, with polymorphism percentage 36.3% (Table 4). The two unique bands at 70.0 and 60.2 kDa characterized R. hybrida only (Figure 4). The limitation of information obtained from biochemical markers may be due to the presence of high contents of polyphenols and polysaccharides that make the extraction of protein difficult (Kaul et al., 2009).
Evaluation of genetic diversity by various molecular systems provided different levels of information that could be important in the management of Rosa germplasm. ISSR markers were more efficient based on better reproducibility and polymorphism percentage. SSR and biochemical markers revealed slight polymorphism within and between Rosa species collected from different plantations. This study needs further confirmation using a large number of accessions and species that can clearly reveal and explain the variability at the species level. However, our results may help in the initiation of intraspecific and interspecific cross-breeding programs for improvement of roses in Ta’if.
The authors have not declared any conflict of interests.
REFERENCES
|
Babaei A, Tabaei-Aghdaei SR, Khosh-Khui M, Omidbaigi R, Naghavi MR, Esselink GD, Smulders MJM (2007). Microsatellite analysis of Damask rose (Rosa damascena Mill.) accessions from various regions in Iran reveals multiple genotypes. BMC Plant Biol. 7(1):12.
Crossref
|
|
|
|
Baydar N, Baydar H, Debener T (2004). Analysis of genetic relationships among Rosa damascena plants grown in Turkey by using AFLP and micro¬satellite markers. J. Biotechnol. 111:263-267.
Crossref
|
|
|
|
Bornet B, Branchard M (2004). Use of ISSR fingerprints to detect microsatellites and genetic diversity in several related Brassica taxa and Arabidopsis thaliana. Hereditas 140:245-248.
Crossref
|
|
|
|
Dafny-Yelin M, Guterman I, Menda N, Ovadis M, Shalit M, Pichersky E, Zamir D, Lewinsohn E, Adam Z, Weiss D, Vainstein A (2005). Flower proteome: changes in protein spectrum during the advanced stages of rose petal development. Planta 222:37-46.
Crossref
|
|
|
|
Doyle JJ, Doyle JL (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19:11-15.
|
|
|
|
Farooq A, Kiani M, Khan MA, Riaz A, Khan AA, Anderson N, Byrne DH (2013). Microsatellite Analysis of Rosa damascena from Pakistan and Iran. Hortic. Environ. Biotechnol. 54(2):141-147.
Crossref
|
|
|
|
Grossi C, Raymond O, Jay M (1997). isozyme polymorphism of Rosa spp. and cultivar identification. Euphytica 98:11-19.
Crossref
|
|
|
|
Heldt WH (1997). A leaf cell consists of several metabolic compartments. Plant Biochemistry and Molecular Biology. Oxford University Press, Oxford.
Crossref
|
|
|
|
Jabbarzadeh Z, Khosh-khui M, Salehi H, Saberivand A (2010). Inter simple sequence repeat (ISSR) markers as reproducible and specific tools for genetic diversity analysis of rose species. Afr. J. Biotechnol. 9(37):6091-6095.
|
|
|
|
Jayasree N, Devi BP, Vijaya N (1998). Somatic embryogenesis and isozymes in rose species Rosa hybrida l. cv. king's ransom. Indian J. Genet. 58(4):449-454.
|
|
|
|
Kaul K, Dhyani D, Sharma RK (2009). Evaluation of DNA extraction methods for RAPD, SSR and AFLP analyses of wild rose species. Floricult. Ornam. Biotechnol. 3(1):25-30
|
|
|
|
Kiani M, Zamani Z, Khalighi A, Fatahi R, Byrne DH (2008). Wide genetic diversity of Rosa damascena Mill. Germplasm in Iran as revealed by RAPD analysis. Sci. Hortic. 115:386-392.
Crossref
|
|
|
|
Laemmli U (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685.
Crossref
|
|
|
|
Mirali N, Aziz R, Nabulsi I (2012). Genetic characterization of Rosa damascene growing in different regions of Syria and its relationship to the quality of the essential oils. Int. J. Med. Arom. Plants 2(1):41-52.
|
|
|
|
Mirzaei L, Rahmani F (2011). Genetic relationships among Rosa species based on random amplified polymorphic DNA (RAPD) markers. Afr. J. Biotechnol. 10(55):11373-11377.
|
|
|
|
Nadeem M, Wang X, Akond M, Awan FS, Khan AI, Riaz A, Younis A (2014). Hybrid identification, morphological evaluation and genetic diversity analysis of Rosa × hybrida by SSR markers. Aust. J. Crop Sci. 8(2):183-190.
|
|
|
|
Rohlf FJ (1998). NTSYSpc: Numerical Taxonomy and Multivariate Analysis System, version 2.02. Exeter Software, New York.
|
|
|
|
Rusanov K, Kovacheva N, Vosman B, Zhang L, Rajapakse S, Atanassov A, Atanassov I (2005). Microsatellite analysis of Rosa damasena Mill. Accessions reveals genetic similarity between genotypes used for rose oil production and old damask rose varieties. Theor. Appl. Genet. 111:804-809.
Crossref
|
|
|
|
Scandalios JC (1964). Tissue-specific allozyme variations in maize. J. Hered. 55:281-285.
Crossref
|
|
|
|
Stafne ET, Clark JR, Weber CA, Graham J, Lewers S (2005). Simple sequence repeat (SSR) markers for genetic mapping of raspberry and blackberry. J. Am. Soc. Hortic. Sci. 130(5):722-728.
|
|
|
|
Stegemann H, Afifiy AMR, Hussein KRF (1985). Cultivar Identification of dates (Phoenix dectylifera) by protein patterns. 2nd International Symposium of Biochemical Approaches to Identification of Cultivars. Braunschweig, West Germany. 44 p.
|
|
|
|
Weeden NF, Wendel JF (1990). Genetics of plant isozymes. In. Soltis, D.E. and P.S. Soltis, (Ed.). Isozymes in plant biology. Chapman and Hall Press, London. pp. 46-72.
|
|
|
|
Wendel JF, Weeden NF (1989). Visualization and interpretation of plant allozymes. In: Soltis, D.E. and P.S. Soltis, (Ed.). Allozymes in plant biology, Advances in plant sciences, series 4. Dioscori¬des Press, Portland, OR. pp. 5-45.
|
|
|
|
Widrlechner MP (1981). History and utilization of Rosa damascena. Econ. Bot. 35:42-58.
Crossref
|
|
|
|
Zhang LH, Byrne DH, Ballard RE, Rajapakse S (2006). Microsatellite marker development in rose and its application in tetraploid mapping. J. Am. Soc. Hortic. Sci. 131(3):380-387.
|