ABSTRACT
Indole-3-acetamide (IAM) pathway was proposed to produce indole-3-acetic acid (IAA). This pathway was proven to be functional in many species of bacteria and expected to operate in plants based on the identification of IAM and IAM-hydrolase (AMI) activity in certain plant species. Available putative AMI sequences from a wide array of monocot and dicot plants were identified and the phylogenetic tree was constructed and analyzed. We identified in this tree, a clade that contained sequences from species across the plant kingdom suggesting that AMI is conserved and may have a primary role in plant growth and development. This clade contained the isolated and well characterized Arabidopsis and Nicotiana AMIs. The preliminary reverse transcriptase polymerase chain reaction (RT-PCR) results for the conserved rice AMI (Os04G02780) showed a transcript for the gene encoded by this enzyme at 1, 7 and 21 days after anthesis (DAF) during the active period of IAA accumulation in developing grains. Based on the accumulated data, we suggested that sequences in this clade are conserved and have an important role in IAA synthesis or other primary processes in plant development.
Key words: Auxin, indole-3-acetic acid, indole-3-acetamide, tryptophan, IAM-hydrolase.
Auxin, from the Greek word "auxein" which means to grow, is found in animals, plants, fungi, and bacteria but having growth regulation effects only in plants. Auxin is an essential plant hormone affecting virtually all aspects of plant growth and development. It affects cell division, elongation, embryogenesis, fruit development, organ patterning, root growth, lateral root initiation, leaf expansion, shoot architecture, and vascular develop-ment. Despite the century-long history of auxin research, the complete picture of how plants synthesize IAA is far from complete understanding. Two major pathways for IAA production have been proposed: the tryptophan (Trp)-dependent pathways and a Trp-independent one. Tryptamine pathway, indole-3-pyruvic acid pathway (IPA), indole-3-acetaldoxime (IAOx) pathway and IAM pathway were proposed to convert Trp to IAA. Plant genes implicated in IAA biosynthesis as well as the reactions catalyzed by the encoded enzymes are illustrated in Figure 1.
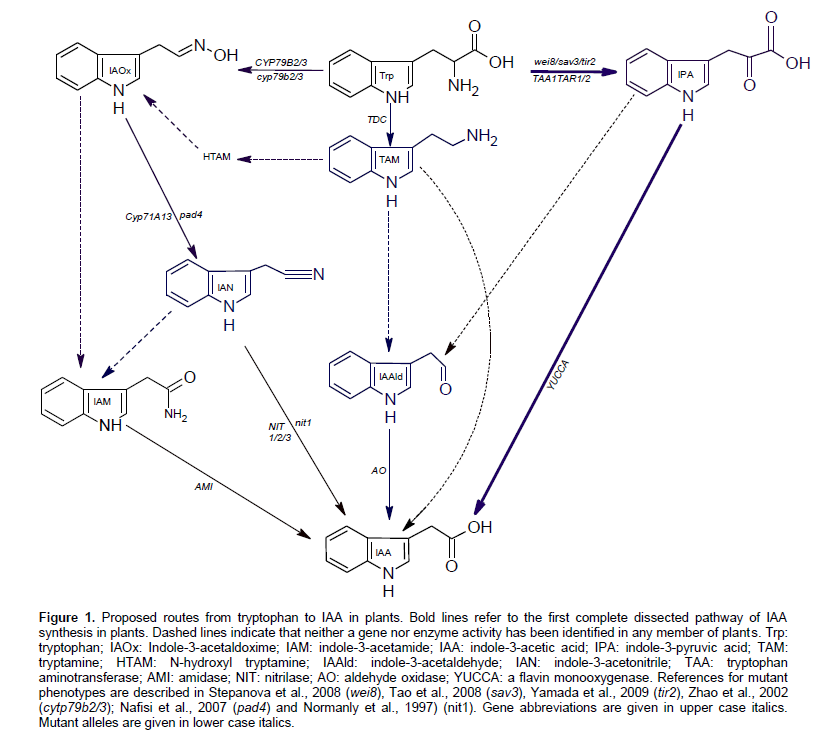
The IPA pathway has recently fully elucidated and proven functional in Arabidopsis (Mashiguchi et al., 2011; Won et al., 2011). In this pathway, a tryptophan amino-transferase (TAA) converts Trp to IPA whereas; YUCCA, A flavin monooxygenase, catalyzes the conversion ofIPA to IAA. IAA produced via IPA pathway was found to have a critical role in various developmental processes including embryogenesis, flower development, seedling growth, vascular patterning, lateral root formation, tropism, shade avoidance, and temperature-dependent hypocotyl elongation (Dai and Zhao, 2006, 2007; Stepanova et al., 2008; Tao et al., 2008).
The mutants in TAA or YUCCA of Arabidopsis are not lethal suggesting that other pathways together with the IPA are expected. The mutant in Vanishing tassel2 (Vt2), which encode grass-specific TAA, was reported to account for 60% reduction in IAA level (Phillips et al., 2011). This hormone in plant is crucial and two pathways of IAA production or more may be needed to ensure the efflux of enough amount of IAA. Moreover, multiple IAA biosynthetic pathways may contribute to regulation of IAA production. Moreover, two pathways of IAA synthesis in bacteria have been fully elucidated.
Indole-3-acetamide has also been proposed to be an IAA precursor in higher plants. The occurrence of IAM as an endogenous compound was reported in young fruits of Citrus unshiu (Takahashi et al., 1975), hypocotyls of Japanese cherry (Saotome et al., 1993), seedlings of A. thaliana, coleoptiles of rice and maize as well as apexes of tobacco (Sugawara et al., 2009). IAM was also detected in a sterile-grown Arabidopsis thaliana and the microbial contributions were found to be insignificant (Pollmann et al., 2002). Pollmann et al. (2009) suggested the role of IAM as an intermediate in IAA synthesis based on the observation that Trp is converted to IAA via IAM in the cell-free Arabidopsis extract.
The enzyme that catalyze the conversion of Trp to IAM is unknown, but the gene encodes IAM-hydrolase which converts IAM to IAA, has been initially cloned and characterized in A. thaliana (Pollmann et al., 2003), and Nicotiana tabacum (Nemoto et al., 2009). IAM-hydro-lyzing activity was also observed in wild and cultivated rice as well as cell-free extracts from various tissues of trifoliata orange (Poncirus trifoliata Rafin) (Kawaguchi et al., 1993; Kawaguchi et al., 1991). AMI was partially purified from rice cells and found to hydrolyze IAM, IAA ethyl ester in addition to the IAM homologue, 1-naph-thalene-acetamide (Arai et al., 2004).
The indole-3-acetamide pathway of auxin biosynthesis has been worked definitely in bacteria. Tryptophan-2-monooxygenase, encoded by the iaam gene, catalyses the first step in this reaction converting Trp to IAM. The second step (IAM to IAA) is catalyzed by IAM-hydrolase that encoded by the iaaH gene. This pathway was found to be operated in a number of bacterial species including Agrobacterium (Inze et al., 1984), Azospirillum (Bar and Okon, 1993), Streptomyces (Manulis et al., 1994), methylobacteria, Bradyrhizobium (Sekine et al., 1989), Rhizobium (Theunis et al., 2004), Pantoea agglomerans (Manulis et al., 1998), and Pseudomonas (Magie et al., 1963).
A high degree of similarity between IAA biosynthesis pathways in plants and bacteria was observed. Trypto-phan has been identified as the main precursor for IAA biosynthesis pathways in bacteria and plants. More than one pathway of IAA synthesis was found to be operated in a single strain of bacteria (Patten and Glick, 1996) and the same thing is expected in plants. IPA was found to be an important intermediate in IAA synthesis in bacteria and a crucial role for this intermediate has been recently confirmed in plants. IAM was also proved functional in bacteria and several lines of evidence for its role in IAA synthesis were found in plants. Differences in IAM reactions between bacterial and plant pathways are also expected.
Because auxin is almost implicated in every step in plant development, enzymes involved in IAA synthesis were expected to be conserved across the plant king-dom. Based on this hypothesis it was expected to find a conserved clade for IAM- hydrolase enzyme proposed to have a role in IAM pathway of IAA synthesis from all studied plant species. Abu-Zaitoon et al. (2012) quantified the free level of IAA in developing rice grains at 1, 4, 7, 10, 14, and 21 DAF using liquid chromatography electrospray ionization / mass spectrometry (LC-ESI-MS). The amount of IAA was found to increase from below the detection limit at 1 DAF to 1.5 µg/gfw at 21 DAF. Rice kernels were therefore used as an experimental material and 1,7,21 DAF were selected to detect the expression profile of AMI.
The major aim of the research described in this work was to discuss in the light of phylogenetic analysis whether the AMI enzyme is conserved across the plant kingdom and therefore its potential importance as an important enzyme in IAA synthesis. Additionally, the involvement of the indole-3-acetamide pathway in IAA production in plants and rice, in particular, will be discussed based on the expression profile of the conserved AMI gene as well as the recent available publications.
Bioinformatics
Bioinformatics was used to investigate the phylogenetic relationships of AMI enzymes proposed to be involved in the IAM pathway of IAA production. IAM-hydrolase from Arabidopsis was used as query sequences. Homologues of the targeted proteins from diverse taxonomic groups of moss, non-seed vascular plants, monocots, and dicots were identified. Constructed phylogenetic tree for IAM-hydrolyase enzymes were then investigated to identify a clade, if any, that are conserved across the plant kingdom.
Homologues of AMI1 were identified following BlastP searches (Altschul et al., 1997) against proteomes of Arabidopsis thaliana, Physcomitrella patens, Selaginella moellendorffii, Oryza sativa subsp. Japonica, Zea mays, Sorghum bicolor, Populus trichocarpa, Vitis vinifera, Glycine max, Fragaria vesca, Medicago truncatula, Ricinus communis, Theobroma cacao, Carica papaya, Nicotiana tabacum, Hordeum vulgare subsp. Vulgare, Brachypodium distachyon, and Picea sitchensis (Plaza 2.5; http://bioinformatics.psb.ugent.be/plaza/ (Bel et al., 2012; Proost et al., 2009), as well as Solanum lycopersicum (The Sol Genomics Network (SGN; http://solgenomics.net/) (Bombarely, et al., 2011). The GenBank database (Non-redundant protein sequences) was screened to search for sequences from other plants. Phylogeny.fr free web service was used to construct and analyze phylogenetic relationships between AMI sequences (Dereeper et al., 2010; Dereeper et al., 2008).
Alignment of amino acids rather than nucleotide sequences was used in this study because it is easier to find unauthentic alignment of DNA sequence which consists of only 4 nucleotides comparing to 20 different amino acids for protein sequences. DNA mutates quickly as a 24% of single base changes produce the same amino acid. Sequences were aligned with MUSCLE (v3.7 with default settings) configured for highest accuracy. Edgar (2004) reported that MUSCLE can achieve both better average accuracy and better speed than CLUSTALW or T?Coffee, depending on the chosen options. Default parameters were claimed to give the best average accuracy.
Distance-matrix (Fitch, 1981), maximum parsimony (Fitch, 1971), and maximum likelihood (Felsenstein, 1981) are the three major methods used to construct phylogenetic trees. Maximum likelihood method implemented in the PhyML program (v3.0 aLRT) (Guindon and Gascuel, 2003) was used to reconstruct the tree. Due to simplicity as well as accuracy and speed, PhyML has been widely used (>2500 citations in ISI Web of Science) (Guindon et al., 2010). Approximate Likelihood-Ratio Test (aLRT) rather than Bootstrapping procedure was used to statistically test branches support. SH-like algorithm was used to estimate the confidence of the aLRT statistics. Default substitution model was used.
MUSCLE (Edgar, 2004) was initially used to perform multiple sequence alignments. The fractional identity between each pair of sequences is computed using k-mer counting. A tree is then con-structed from the triangular distance matrix computed from the pair-wise similarities using UPGMA and a root is identified. The resulting tree is improved by building a new progressive alignment and a new tree is constructed by computing a Kimura distance matrix. SH-like is an algorithm used to estimate the confidence of the aLRT statistics. This method of branch test is considered to be a very fast, alternative and complement to the standard bootstrap analysis. An appropriate array is stored from simply drawn and summed values. Moreover, the nonparametric SH-like support is closely related to P values of statistical tests. Experiments with simulated data indicate that the new SH-like interpretation of the aLRT statistic should be preferred to the parametric chi-square–based interpretation due to unavoidable simplifications of substitution models when analyzing real data. As the standard bootstrap, aLRT with SH-like interpretation is conservative (Guindon et al., 2010).
Plant materials
Rice plants, Oryza sativa (c.v. Jarrah) were grown in a glasshouse in flooded soil-filled plastic pots. Pots were filled with black soil containing Aquasol fertilizers (N: P: K 23:4:18). Temperature inside the green house was 30°C during the day and 18°C during the night. Each branch of rice plants was marked when the top half of the panicle had reached anthesis. Superior caryopses were harvested 1, 7, and 21 DAF. Microfuge tubes containing 60 to 70 mg of rice seed (10, 3, and 2 seeds from 1, 7, and 21 day samples respectively) were then weighed and frozen in liquid nitrogen for total RNA extraction. Samples not used immediately were stored in a freezer at -70°C for later use.
RNA Isolation
Total RNA from rice grains was extracted following the instructions for RNeasy® Mini Kit (Qiagen, Cat. No. 74903). The absorbance of extracted RNA was measured at 260 nm (A 260). The con-centration of RNA in µg/mL was calculated as follows: Concen-tration= 40* A 260* Dilution Factor (QIAGEN, 2002). A nano-drop spectrophotometer (NanoDrop® ND-1000 Spectro-photometer, NanoDrop Technologies, Inc., USA) was also used to measure RNA concentration, A260/A280 and A260/A230 ratios. RNA extracts with high purity, A260/A280 and A260/A230 > 1.8 and 2 respectively, were used in all experiments (QIAGEN manual). RNA quality was checked by electrophoresis in 1.2% agarose run at 100V for 45 min after heating at 80 ?C for 10 min and cooling on ice (Nolan et al., 2006).
Polymerase chain reaction
The program, primer3 (Rozen, 1998) was used to design primers for the cDNAs of all targeted genes. Primer sequences are listed in Table 1. Gene-specific primers were designed so that either the left or right primer was complementary to an exon-exon boundary to exclude amplification of genomic DNA. Primers were dissolved in fresh sterile milli-Q water to prepare 50 µM stock solution; 10 µM working solutions were prepared and frozen at -20 °C.
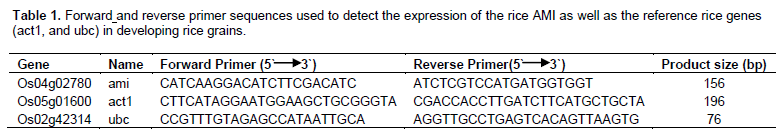
The reverse transcriptase polymerase chain reaction with gene-specific primers was run according to the instructions of the manufacturer (QIAGEN®, OneStep RT-PCR Kit, Lot No. 133206209). Thermal cycler (PTC-100 TM Programmable Thermal Controller, MJ Research, Inc., USA) conditions are listed in Table 1. The same amount of RNA extracts (1.2 µg) was used to perform all RT-PCR reactions. 3 µL each of 10 µM primer solution was added to a final concentration of 0.6 µM. RT-PCR was attempted using 30 cycles. To estimate the size of each RT-PCR product, BenchTop 100 bp DNA Ladder was used (Promega). The intensity of the 500 band is three times higher than that of the other equal intensity bands and as a result can be used as a reference band.
Purification and sequencing
For PCR, the amplification protocol was the same as indicated for RT-PCR but with 35 cycles and without the reverse transcriptase reaction. cDNA products were purified according to the Wizard SV Gel and PCR Clean-Up System (Promega, Cat. # A9281). Concen-tration of cDNA samples was calculated according to the following equation Concentration= 50* A 260* Dilution Factor (Sambrook and Russell, 2001). Full-length cDNA of all genes were sequenced by Prince Alfred Molecular Analysis Centre, Sydney University.
Phylogenetic relationships of IAM-hydrolase from the available sequenced plant genomes were investigated. Arabidopsis AMI1 (AtAMI1) was used as a query sequence to search for all putative AMI in plants. As a result of BlastP search using the available plant proteomic data bases, 154 putative AMI sequences were found. The number of sequences ranges from 1 for P. sitchensis to 21 for P. trichocarpa. To simplify the phylogenetic tree, sequences from the same species that are much more similar to each other rather than to sequences from other plants were omitted. Therefore, the number of sequences was reduced to 138.The phylogenetic tree for the 138 IAM hydrolase sequences (figure not shown due to the large number of sequences) showed that all sequences are classified into two major branches. The majority of plant sequences are found in the first major branch that is supported at 88% by statistical analysis. For example O. sativa, Z. maysand S. bicolor have 9, 6 and 7 sequences, respectively in this branch. The Arabidopsis sequence (At5g64440) that was not found to hydrolyze indole-3-acetamide detectably is classified in the first branch suggesting that sequences in this branch may not be involved in IAA synthesis. At5g64440, a fatty acid amide hydrolase (Shrestha et al., 2003), has the ability to convert N-acylethanolamine to ethanolamine and the corresponding free fatty acids. Large amount of ester IAA for plants including rice may explain the large number of putative amidases in this branch. Enzyme kinetics and substrate specificity of the other two Arabidopsis sequences (At5G07360, At3G25660) in this clade needs to be worked out. However, these two sequences have the essential residues of the supposed amidase activity.
To get a more precise picture, phylogenetic tree for all sequences in the second major branch was reconstructed at high stringent selection (Figure 2). Unlike the first major branch, all studied plants were found to have representative sequences in this branch. The second major branch that is supported at 100% by statistical analysis is divided into two minor branches. The first minor one, supported at 94% by statistical analysis, contains the Arabidopsis sequence (At5G09420) that does not found to have amidase activity suggesting that representatives of this branch may not be involved in IAA synthesis. At5G09420 is localized in the outer mitochondrial membrane in complex with a protein translocase (Chew et al., 2004).
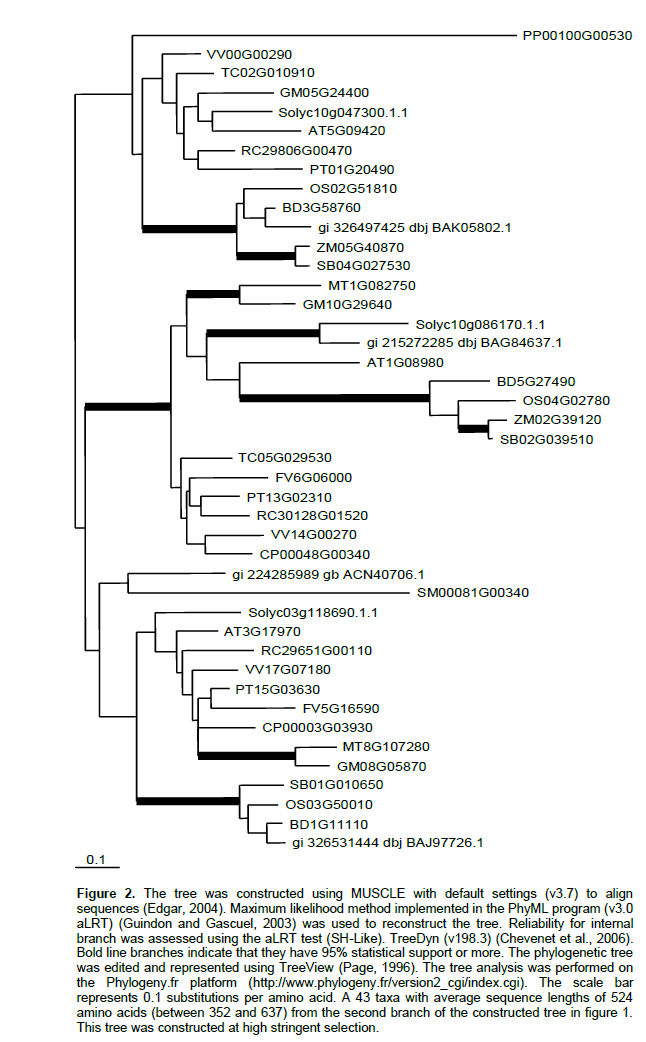
The second minor branch, supported at 94% by statistical analysis, is also divided into two clades. The well characterized Arabidopsis amidase 1(AMI1) and N. tabacum 1 (NtAMI) as well as the only two sequences of H. vulgare are found in the first clade that was supported at 100% by statistical analysis. The only sequence of P. sitchensis is also found in this clade. No any sequences for P. patens and S. moellendorffii were identified in this clade. Unlike other Arabidopsis putative amidases that was not found to be involved in IAA synthesis and found in mitochondria or chloroplast, the intracellular location of the AtAMI1 was identified in the cytoplasm where IAA synthesis occurs (Pollmann et al., 2006). All sequences except Pt13G02310 in this clade contains the essential residues of the supposed amidase catalytic triad (Lys-cisSer-Ser) (Shin et al., 2003) (Figure 3). The first clade is conserved and expected to have a role in IAA synthesis or other primary role in plant growth and development. Characterization of amidases in this clade from different plants especially those of agricultural importance that are generally used in IAA studies; i.e rice, maize, and tomato, will gather pieces of knowledge to form a complete picture of IAM in plants.

The second clade, supported at 74% by statistical analysis, contains the Arabidopsis sequence (At3G17970) which is not expected to have amidase activity. At3G17970, a part of the preprotein translocon of the outer envelope of chloroplasts (Toc64-III), shows some structural differences from AtMI1. Toc64-III contains tetratricopeptide repeat motifs in its C-terminal domain that are found in the ancient moss P. patens (Hofmann and Theg, 2003) but not the essential residues of the supposed amidase catalytic triad.
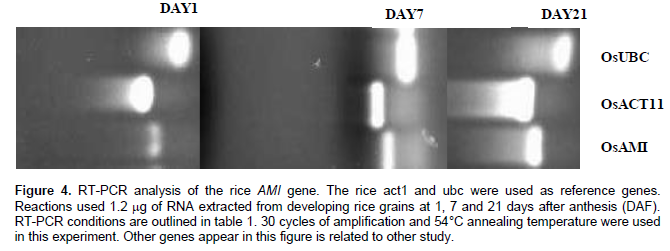
IAM hydrolase was partially purified from rice cells and reported to hydrolyze IAA ethyl ester as well as IAM and its homologue 1-naphthalene acetamide but not indole-3-acetonitrile (Arai et al., 2004). Although the molecular mass of the enzyme was very similar to that of the Agrobacterium tumefaciens, a significant role for this enzyme in IAA synthesis was excluded due to the considerably high km value. AMI generally has dual functions as amidase and esterase. Together with the observation that rice ears contain a large amount of esterified IAA at the stage of anthesis (Kobayashi et al., 1989). Arai et al. (2004) suggested that this enzyme may serve to control IAA accumulation via the hydrolysis of IAA. In this study we reported that rice have 15 putative IAM hydrolase enzymes, one of them (Os04G02780) is in the conserved clade. As a preliminary result, a transcript for Os04G02780 was detected in rice grain at 1, 7 and 21 days after anthesis where large amount of IAA is accumulated during this period of growth (Figure 4). Therefore, it is expected for Os04G02780 to have a significant role in IAA synthesis via the IAM intermediate.
Pollmann et al. (2009) isolated IAA synthase, an enzyme complex that can convert Trp to IAA, from 14 different plant species. They suggested that IAM is an intermediate in IAA synthesis by this complex due to the finding that IAM interferes with the conversion of 1mM [2H]5-Trp to [2H]5- IAA. The reduction in IAA conversion was reported to b 66% and 80% in case of 1 mM and 0.1 mM IAM respectively. IAA synthase was also reported to convert labeled IAM to IAA and increasing concentrations of Trp interfered with this conversion. The amount of IAM in Arabidopsis seedlings was found at levels similar to that of free IAA (Pollmann et al., 2002). Based on the higher conversion rate of L-Trp and [2H]5-IAM to IAA comparing to excess L-Trp, Pollmann et al. (2009) concluded that the formation of IAM rather than the conversion of IAM to IAA is the rate limiting step in IAM pathway of IAA synthesis.
The physiological importance of the IAM pathway of IAA synthesis came from the observation that transgenic BY-2 cells overexpressing N. tabacum AMI1 could grow in medium containing low concentrations of IAM, whereas suppression of the NtAMI1 gene by RNA interference caused severe growth inhibition in medium containing IAM. These results clearly show that the NtAMI1 gene is expressed in plant cells and is required for the conversion of IAM to the auxin IAA that involved in plant cell division (Nemoto et al., 2009).
The same scenario as IPA pathway in bacteria and plants, the possibility for the direct conversion of Trp to IAM and therefore the 2-step IAM pathway of IAA synthesis in plant as bacteria is not ruled out. Generatinga mutant with a decreased level of IAM will help to find out the enzyme that catalyses the rate limiting step of this pathway. Making a microarray screens for the expression level of plant genes before and after silencing IAM hydrolase may give us some indications about the change in the gene level for the enzyme catalyze the missed step in the IAM pathway.
The author(s) have not declared any conflict of interests.
REFERENCES
Abu-Zaitoon YM, Bennett K, Normanly J, Nonhebel HM (2012). A large increase in IAA during development of rice grains correlates with the expression of tryptophan aminotransferase OsTAR1 and a grain-specific YUCCA. Physiol. Plant. 146(4):487-499.
Crossref |
|
|
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402.
Crossref |
|
|
Arai Y, Kawaguchi M, Syono K, Ikuta A (2004). Partial purification of an enzyme hydrolyzing indole-3-acetamide from rice cells. J. Plant Res. 117:191-198.
Crossref |
|
|
Bar T, Okon Y (1993). Tryptophan conversion to indole-3-acetic-acid via indole-3-acetamide in Azospirillum-brasilense Sp7. Can. J. Microbiol. 39(1):81-86.
Crossref |
|
|
Bel MV, Proost S, Wischnitzki E, Movahedi S, Scheerlinck C, Peer YVd, Vandepoele K (2012). Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 158:590-600.
Crossref |
|
|
Bombarely A, Menda N, Tecle I, Buels R, Strickler S, Fischer-York T, et al. (2011). The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res. 39:D1149-D1155
Crossref |
|
|
Chevenet F, Brun C, Banuls AL, Jacq B, Chisten R (2006). TreeDyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7:439.
Crossref |
|
|
Chew O, Lister R, Qbadou S, Heazlewood J, Soll J, Schleiff E, et al. (2004). A plant outer mitochondrial membrane protein with high amino acid sequence identity to a chloroplast protein import receptor. FEBS Lett. 557(1-3):109-114.
Crossref |
|
|
Dereeper A, Audic S, Claverie JM, GB (2010). BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10:8.
Crossref |
|
|
|
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Web Server issue):W465-469. |
|
|
Edgar RC (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113.
Crossref |
|
|
Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792-1797.
Crossref |
|
|
Felsenstein J (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17(6):368-376.
Crossref |
|
|
Fitch WM (1971). Toward defining the course of evolution - minimum change for a specific tree topology. Syst. Zool. (20):406-416.
Crossref |
|
|
Fitch WM (1981). A non-sequential method for constructing trees and hierarchical classifications. J. Mol. Evol. 18(1):30-37.
Crossref |
|
|
Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010). New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 59(3):307-321.
Crossref |
|
|
Guindon S, Gascuel OA (2003). simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52(5):696-704.
Crossref |
|
|
Hofmann NR, Theg SM (2003). Physcomitrella patens as a model for the study of chloroplast protein transport: conserved machineries between vascular and non-vascular plants. Plant Mol. Biol. 53(5): 643-654
Crossref |
|
|
Inze D, Follin A, Van Lijsebettens M, Simoens C, Genetello C, Van Montagu M, Schell J (1984). Genetic analysis of the individual T-DNA genes of Agrobacterium tumefaciens; further evidence that two genes are involved in indole-3-acetic acid synthesis. Mol. Gen. Genet. 194(1):265-274.
Crossref |
|
|
|
Kawaguchi M, Fujioka S, Sakurai A, Yamaki YT, Syono K (1993). Presence of a Pathway for the Biosynthesis of Auxin via Indole-3-Acetamide in Trifoliata Orange. Plant Cell Physiol. 34(1):121-128. . |
|
|
|
Kawaguchi M, Kobayashi M, Sakurai A, Syono K (1991). The Presence of an Enzyme that Converts IndoIe-3-acetamide into IAA in Wild and Cultivated Rice. Plant Cell Physiol. 32(2):143-149. |
|
|
Kobayashi M, Sakurai A, Saka H, Takahashi N (1989). Fluctuation of the endogenous IAA level in rice during its life cycle. Agric. Biol. Chem. 53:1089-1094.
Crossref |
|
|
Magie AR, Wilson EE, Kosuge T (1963). Indoleacetamide as an Intermediate in the Synthesis of Indoleacetic Acid in Pseudomonas savastanoi. Science 141(3587): 1281-1282.
Crossref |
|
|
Manulis S, Haviv-Chesner A, Brandl M, Lindow S, Barash I (1998). Differential involvement of indole-3-acetic acid biosynthetic pathways in pathogenicity and epiphytic fitness of Erwinia herbicola pv. gypsophilae. Mol. Plant Microbe Interact. 11:634-642.
Crossref |
|
|
Manulis S, Shafrir H, Epstein E, Lichter A, Barash I (1994). Biosynthesis of indole-3-acetic acid via the indole-3-acetamide pathway in Streptomyces spp. Microbiology 140(5):1045-1050.
Crossref |
|
|
Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. PNAS 108(45):18512-18517.
Crossref |
|
|
Nafisi M, Goregaoker S, Botanga CJ, Glawischnig E, Olsen CE, Halkier BA, Glazebrook J (2007). Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19(6):2039-2052.
Crossref |
|
|
Nemoto K, Hara M, Suzuki M, Seki H, Muranaka T, Mano Y (2009). The NtAMI1 gene functions in cell division of tobacco BY-2 cells in the presence of indole-3-acetamide. FEBS Lett. 583:487-492.
Crossref |
|
|
Nolan T, Hands RE, Bustin SA (2006). Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1(3):1559-1582.
Crossref |
|
|
Normanly J, Grisafi P, Fink G, Bartel B (1997). Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 9(10):1781-1790.
Crossref |
|
|
|
Page RDM (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. |
|
|
Patten CL, Glick BR (1996). Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 42(3):207-220.
Crossref |
|
|
Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C (2011). Vanishing tassel2 Encodes a Grass-Specific Tryptophan Aminotransferase Required for Vegetative and Reproductive Development in Maize. Plant Cell 23:550-566
Crossref |
|
|
Pollmann S, Düchting P, Weiler E (2009). Tryptophan-dependent indole-3-acetic acid biosynthesis by IAA-synthase proceeds via indole-3-acetamide. Phytochemistry 70(4):523-531.
Crossref |
|
|
Pollmann S, Muller A, Piotrowski M, Weiler EW (2002). Occurrence and formation of indole-3-acetamide in Arabidopsis thaliana. Planta 216(1):155-161.
Crossref |
|
|
Pollmann S, Neu D, Lehmann T, Berkowitz O, Schafer T, Weiler EW (2006). Subcellular localization and tissue specific expression of amidase 1 from Arabidopsis thaliana. Planta 224:1241-1253.
Crossref |
|
|
Pollmann S, Neu D, Weiler E (2003) . Molecular cloning and characterization of an amidase from Arabidopsis thaliana capable of converting indole-3-acetamide into the plant growth hormone, indole-3-acetic acid. Phytochemistry 62(3):293-300.
Crossref |
|
|
Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K (2009). PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell 21: 3718-3731.
Crossref |
|
|
|
QIAGEN (2002). QIAGEN OneStep RT-PCR Kit Handbook. |
|
|
|
Rozen SaS HJ (1998). Primer3, from
View
|
|
|
|
Sambrook J, Russell DW (2001). Preparation and Analysis of Eukaryotic Genomic DNA (third edition ed. Vol. 1). New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor. |
|
|
|
Saotome M, Shirahata K, Nishimura R, Yahaba M, Kawaguchi M, Syono K (1993). The Identification of Indole-3-Acetic Acid and Indole-3-Acetamide in the Hypocotyls of Japanese Cherry. Plant Cell Physiol. 34(1):157-159. |
|
|
|
Sekine M, Watanabe K, Syono K (1989). Molecular Cloning of a Gene for Indole-3-Acetamide Hydrolase from Bradyrhizobium japonicum. J. Bacteriol. 171(3):1718-1724. |
|
|
Shin S, Yun Y, Koo H, Kim Y, Choi K, Oh B (2003). Characterization of a novel Ser-cisSer-Lys catalytic triad in comparison with the classical Ser-His-Asp triad. J. Biol. Chem. 278(27):24937-24943.
Crossref |
|
|
Shrestha R, Dixon RA, Chapman KD (2003). Molecular Identification of a Functional Homologue of the Mammalian Fatty Acid Amide Hydrolase in Arabidopsis thaliana. J. Biol. Chem. 278: 34990-34997.
Crossref |
|
|
Stepanova A, Robertson-Hoyt J, Yun J, Benavente L, Xie D, Dolezal K (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133(1):177-191.
Crossref |
|
|
Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Koshiba T(2009). Biochemical analyses of indole-3-acetaldoximedependent auxin biosynthesis in Arabidopsis. P. Natl. A. Acad. Sci. USA. 106(13):5430-5435.
Crossref |
|
|
|
Takahashi N, Yamaguchi I, Kono T, Igoshil M, Hirose K, Suzuki K (1975). Characterization of plant growth substances in Citrus unshiu and their change in fruit development. Plant Cell Physiol. 16:1101-1111. |
|
|
Tao Y, Ferrer J, Ljung K, Pojer F, Hong F, Long J (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133(1):164-176.
Crossref |
|
|
Theunis M, Kobayashi H, Broughton WJ, Prinsen E (2004). Flavonoids, NodD1, NodD2, and Nod-Box NB15 Modulate Expression of the y4wEFG Locus That Is Required for Indole-3-Acetic Acid Synthesis in Rhizobium sp. strain NGR234. Mol. Plant Microbe Interact. 17(10): 1153-1161.
Crossref |
|
|
Won C, Shen X, Mashiguchi K, Zheng Z, Daia X, Cheng Y (2011). Conversion of tryptophan to indole-3-acetic acid by tryptophan aminotransferases of Arabidopsis and YUCCAs in Arabidopsis. PNAS 108(45):18518-18523.
Crossref |
|
|
Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M (2009). The TRANSPORT INHIBITOR RESPONSE2 Gene Is Required for Auxin Synthesis and Diverse Aspects of Plant Development. Plant Physiol. 151:168-179.
Crossref |
|
|
Zhao YD, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR (2002). Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16(23):3100-3112.
Crossref |