Full Length Research Paper
ABSTRACT
Jatropha curcas is an interesting alternative for biodiesel production due to the high oil content in its seeds, its ability to grow in a wide range of climate and soil conditions as well as low cost of production. However, the species is considered to be in domestication and there are no defined cultivars. Therefore, it is extremely important to understand the genetic diversity of the species for selection and characterization of promising genotypes to initiate breeding programs. The objective of this study is to evaluate the phenotypic diversity of physic nut in order to select the most divergent and superior genotypes to compose future breeding programs, using multivariate analysis. Eleven agronomic characters were evaluated in 165 J. curcas genotypes from the in vivo germplasm bank, which were: Plant height, stem diameter, number of primary branches, fruit length, width, weight and shape, seed length, width and weight plus the oil content. The data were analyzed by principal component analysis (PCA), cluster analysis by Ward and k-means methods. The character fruit shape was removed from the multivariate analysis as the only one with qualitative character. The PCA resulted in 4 main components (PC), which explained 71.62% of total variance. The characters selected in PC1 were seed weight, fruit width, fruit length and fruit weight. There were 22 promising genotypes highlighted, with potential to be exploited in breeding programs. Cluster analysis by Ward and k-means methods generated 9 groups influenced by all analyzed characters, of which five groups of genotypes had advantageous characters. Regarding fruit shape, 13 genotypes had an ellipsoid lanceolate shape and the others had an ellipsoid spherical shape. Multivariate analyses allowed genotype characterization, indicating good strategies used for the selection in genetic breeding programs.
Key words: Agronomic characters, Jatropha curcas, oleaginous, multivariate analysis.
INTRODUCTION
MATERIALS AND METHODS
RESULTS AND DISCUSSION
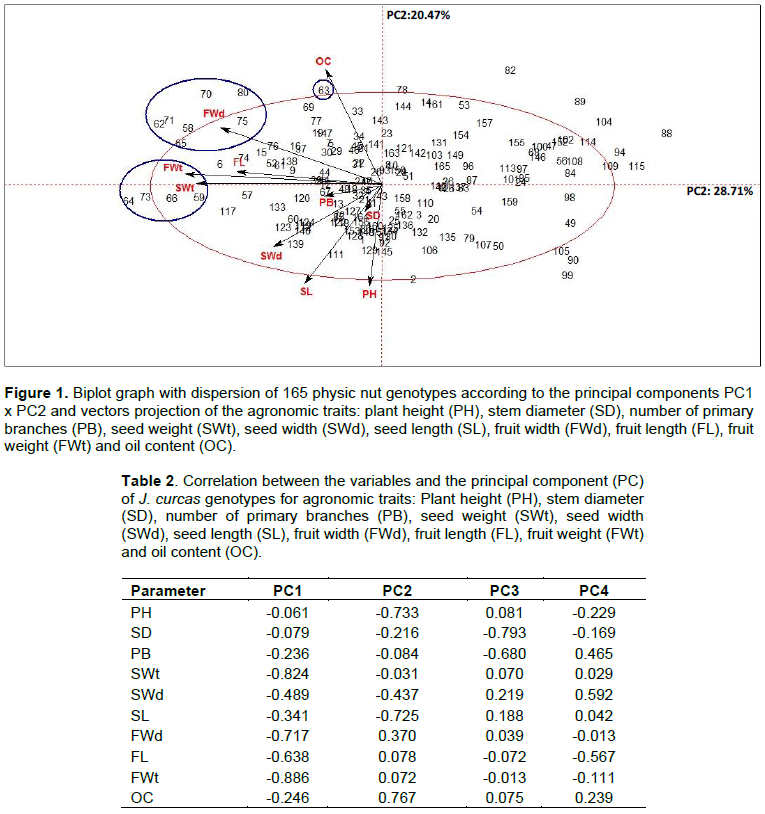
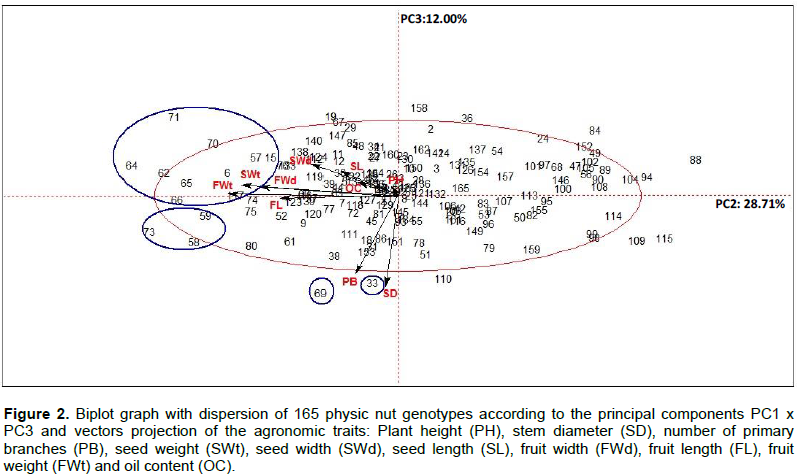
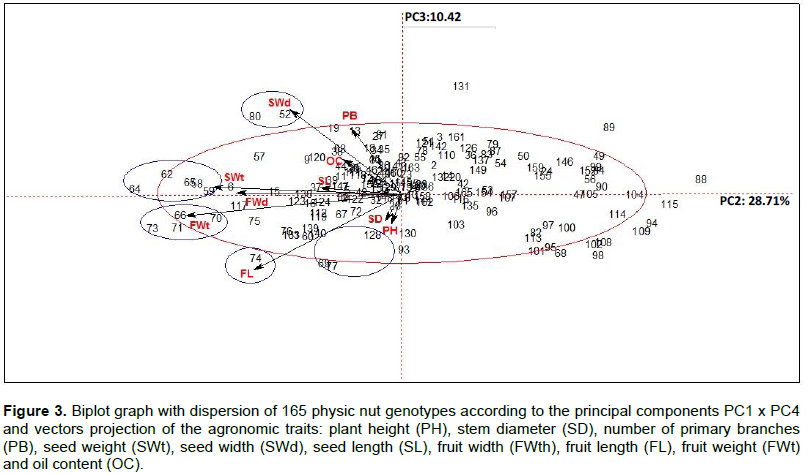
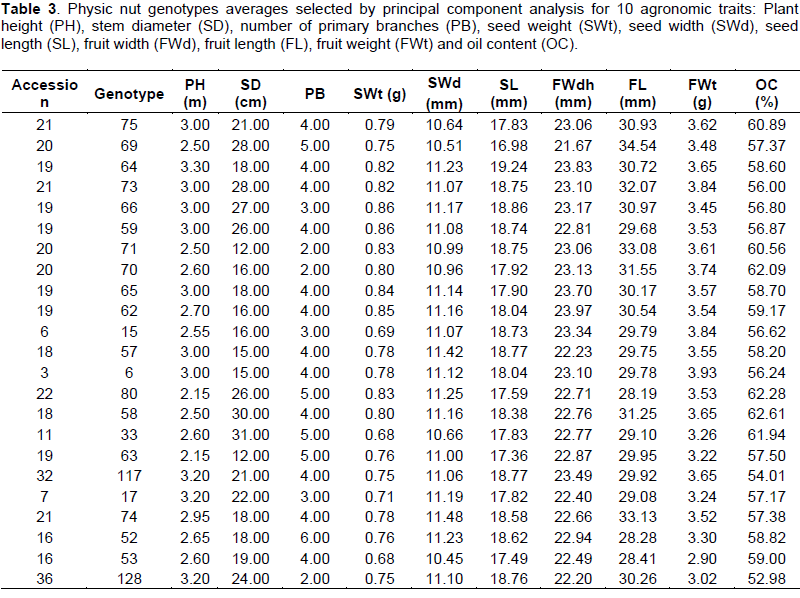
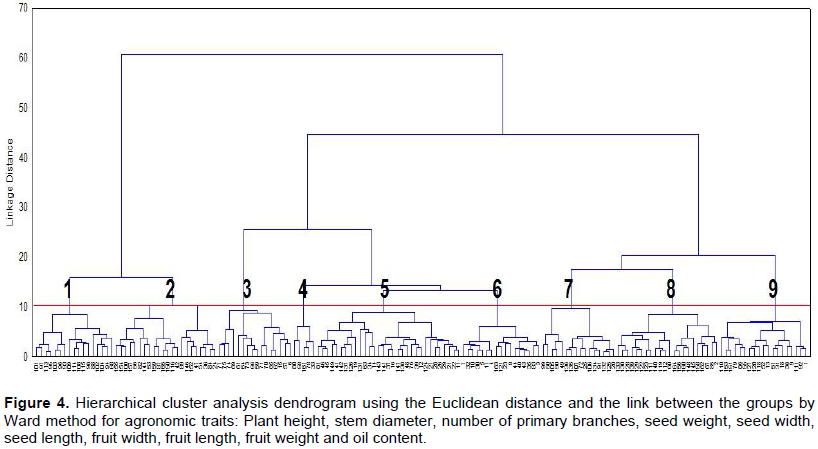
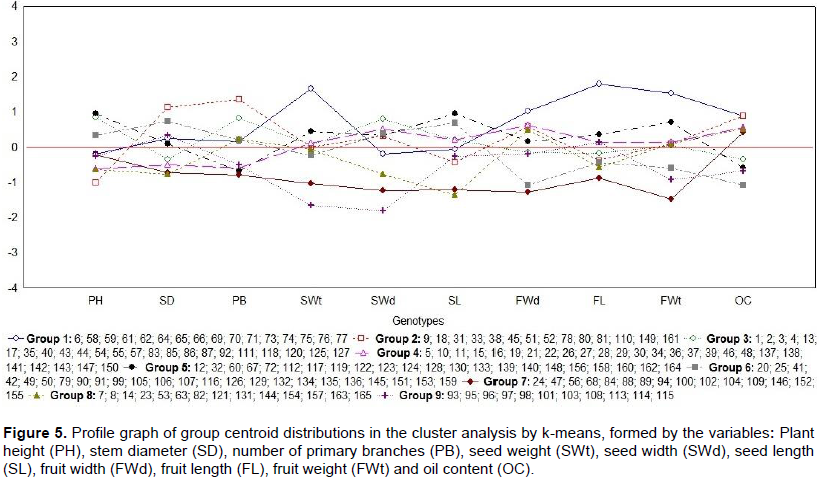
CONCLUSIONS
CONFLICT OF INTERESTS
REFERENCES
|
Abreu FB, Resende MDV, Anselmo JL, Saturnino HM, Brenha JAM, Freitas, FB (2009). Variabilidade genética entre acessos de pinhão-manso na fase juvenil. Magistra 21(1):36-40. |
|
|
Achten WMJ, Nielsen LR, Aerts R, Lengkeek AG, Kjaer ED, Trabucco A, Hansen JK, Maes WH, Graudal L, Akinnifesi FK, Muys B (2010). Towards domestication of Jatropha curcas. Biofuel 1(1):91-107. |
|
|
Achten WMJ, Verchot L, Franken YJ, Mathijs E, Sing VP, Aerts R, Muys B (2008). Jatropha bio-diesel production and use. Biomass Bioenergy 32(12):1063-1084. |
|
|
Aguilera-Cauich EA, Pérez-Brito D, Yabur AN, López-Puc G, Najera GC, Rivero JCS, Atoche CR, Uc-Várguez A, Góngora-Canul C, Mijangos-Cortes JO (2015). Assessment of phenotypic diversity and agronomic contrast in American accessions of Jatropha curcas L. Ind. Crops Prod. 77:1001-1003. |
|
|
Anjani K (2010). Pattern of genetic diversity among Fusarium wilt resistant castor germplasm accessions (Ricinus communis L.). Elect. J. Plant Breed. 1(2):182-187. |
|
|
AOCS (2003). Official methods and recommended practices of the American Oil Chemistry Society. 5th ed. Champaign. |
|
|
Brasileiro BP, Silva SA, Souza DR, Santos PA, Oliveira RS, Lyra DH (2013). Genetic diversity and selection gain in the physic nut (Jatropha curcas). Genet. Mol. Res.12(3):2341-2350. |
|
|
Carvalho DS (2010). Comportamento genético de progênies de meio-irmãos de pinhão manso no recôncavo baiano, Brasil. Thesis (Master's degree) - Universidade Federal do Recôncavo da Bahia, Cruz das Almas, 51 p. |
|
|
Cruz CD, Regazzi AJ, Carneiro PCS (2004). Modelos biométricos aplicados ao melhoramento genético. In. Cruz CD, Regazzi, JA, Carneiro PCS (Eds.), Viçosa. pp. 377-413. |
|
|
Drummond OA, Purcino AAC, Cunha LHS, Veloso JM (1984). Cultura do pinhão-manso. Belo Horizonte: EPAMIG. 131:99. |
|
|
Eriksson O (1999). Seed size variation and its effect on germination and seedling performance in the clonal herb Canvallaria majalis. Acta Oecol. 20:61-66. |
|
|
Fairless D (2007). Biofuel: the little shrub that could–maybe. Nature 449:652-655. |
|
|
Freitas RG, DIAS LAS, Cardoso PMR, Evaristo AB, Silva MF, Araújo NM (2015). Diversity genetic parameter estimates for yield and its components in Jatropha curcas L. Genet. Mol. Res. 1:1-10. |
|
|
Gonçalves LSA, Rodrigues R, Amaral ATJ, Karasawa M, Sudré CP (2008). Comparison of multivariate statistical algorithms to cluster tomato heirloom accessions. Genet. Mol. Res. 7(4):1289-1297. |
|
|
Iqbal Z, Arshad M, Mahmood T, Waheed A (2008). Evaluation of soybean [Glycine max (L.) Merrill] germplasm for some important morphological traits using multivariate analysis. Pak. J. Bot. 40(6):2323-2328. |
|
|
Jun-Ling S, Xiang-Nan J, Hui-Qun N, Pei-Guang S, Shi-Hui N, Xiao-Yang C (2010). AFLP analysis of genetic diversity of Jatropha curcas grown in Hainan, China. Trees 24(3):455-462. |
|
|
Kaushik N, Kumar K, Kumar S, Kaushik N, Roy S (2007). Genetic variability and divergence studies in seed traits and oil content of Jatropha (Jatropha curcas L.) accessions. Biomass Bioenergy 31:497-502. |
|
|
Laviola BG, Dias LAS (2008). Teor e acúmulo de nutrientes em folhas e frutos de pinhão-manso. Rev. Bras. Ciênc. Solo 32:1969-1975. |
|
|
Laviola BG, Bhering LL, Mendonça S, Rosado TB, Albrecht JC (2011). Caracterização morfo-agronômica do banco de germoplasma de pinhão-manso na fase jovem. Biosci. J. 27(3):371-379. |
|
|
Lima RLS, Severino LS, Sampaio LR, Freire MAO, Beltrão, NEM, Arriel NHC (2009). Crescimento e teor foliar de nutrientes em mudas de pinhão-manso (Jatropha curcas L.) em substratos contendo cinco materiais orgânicos e fertilizante mineral. Rev. Bras. Ol. Fibros. 13(1):29-36. |
|
|
MILANI M (2008). Descritores de mamona utilizados pela Embrapa Algodão. Embrapa Algodão. Documentos. |
|
|
Neto JMM, Moita GC (1998). Uma introdução à análise exploratória de dados multivariados. Quím. Nova 21(4):467-469. |
|
|
Nietsche S, Vendrame WA, Crane JH, Pereira MC, Costa A, Reis ST, (2015). Variability in reproductive traits in Jatropha curcas L. accessions during early developmental stages under warm subtropical conditions. Glob. Change Biol. Bioenergy 7:122-134. |
|
|
Noor Camellia NA, Thohirah Lee A, Abdullah NAP (2012). Genetic relationships and diversity of Jatropha curcas accessions in Malaysia. Afr. J. Biotechnol. 11(13):3048-3054. |
|
|
Oliveira JPM, Silva JF, Oliveira BS, Santos PGF, Silveira PS, Matos FS (2016). Phenotypic characterization of physic nut populations. Afr. J. Agric. Res. 11(45):4559-4566. |
|
|
Oliveira MM, Rota EL, Dionello NJL, Aita MF (2007). Herdabilidade e correlações genéticas do perímetro escrotal com características produtivas em bovinos de corte: Revisão. R. Bras. Agrociência 13:141-146. |
|
|
Pazeto MSR, Unêda-Trevisoli SH, Corrêa AAP, Vianna VF, Leite DC, Di Mauro AO (2015). Genetic diversity in Jatropha species from different regions of Brazil based on morphological characters and inters-simple sequence repeat (ISSR) molecular markers. Afr. J. Biotechnol. 14(25):2066-2079. |
|
|
Pecina-Quintero V, Anaya-López JL, Zamarripa-Colmenero A, Nú-ez-Colín CA, Montes-García N, Solís-Bonilla JL, Jiménez-Becerril MF (2014). Genetic structure of Jatropha curcas L. in Mexico and probable centre of origin. Biomass Bioenergy 60:147-155. |
|
|
Pereira FHF, Puiatti M, Miranda GV, Silva DJH, Finger FL (2003). Divergência genética entre acessos de taro utilizando caracteres morfoqualitativos de inflorescência. Hortic. Bras. 21(3):520-524. |
|
|
Pitta MR, Boiça Jr AL, Jesus FG, Tagliari SRA (2010). Seleção de genótipos resistentes de amendoinzeiro à Anticarsia gemmatalis hübner (Lepidoptera: Noctuidae) com base em análises multivariadas. Neotrop. Entomol. 39(2):260-265. |
|
|
Priyanka, Ram D, Shiva N, Anurag K, Dalbeer (2015). Assessment of variability parameters and character association for quantitative traits in Jatropha. Trends Biosci. 8(15):3896-3899. |
|
|
Rao GR, Korwar GR, Shanker AK, Ramakrishma YS (2008). Genetic associations, variability and diversity in seed characters growth, reproductive phenology and yield in Jatropha curcas L. accessions. Trees 22(5):697-709. |
|
|
Reddi KR, Satya AK, Ramesh P, Lekkala SP, Narendra K, Reddy PCO, Reddy CVCM, Sekhar AC (2016). Molecular genetic assessment of Jatropha curcas L. germplasm of diverse origin along with its wild relatives for various early growth and establishment related traits. Int. J. Pharma Bio Sci. 7(4):(B)261-271. |
|
|
Reis MVM, Junior PCD, Campos TO, Diegues IP, Freitas, SC (2015). Variabilidade genética e associação entre caracteres em germoplasma de pinhão-manso (Jatropha curcas L.). Rev. Ciênc. Agron. 46(2):412-420. |
|
|
Saturnino HM, Pacheco DD, Kakina J, Tominaga N, Gonçalves NP (2005). Cultivation of Jatropha curcas L. Rev. Inf. Agropecuário 26(229):44-78. |
|
|
Shabanimofrad M, Rafii MY, Megat Wahab PE, Biabani AR, Latif MA (2013). Phenotypic, genotypic and genetic divergence found in 48 newly collected Malaysian accessions of Jatropha curcas L. Ind. Crops Prod. 42:543-551. |
|
|
Silva Junqueira SV, Peixoto ALD, Laviola GB, Bhering LL, Mendonça S (2016). Bayesian multi-trait analysis reveals a useful tool to increase oil concentration and to decrease toxicity in Jatropha curcas L. Plos One 11(6):1-10. |
|
|
Singh S, Prakash A, Chakraborty NR, Wheeler C, Wheeler PK, Ghosh A (2016). Trait selection by path and principal component analysis in Jatropha curcas for enhanced oil yield. Ind. Crops Prod. 86:173-179. |
|
|
Sinha P, Islam MDA, Negi MS, Tripathi SB (2016). First identification of core accessions of Jatropha curcas from India based on molecular genetic diversity. Plant Genet. Resour. 14(1):77-80. |
|
|
Spinelli VM, Rocha RB, Ramalho AR, Marcolan AL, Vieira JR, Fernandes CF, Militão JSLT, Dias LAS (2010). Primary and secondary yield components of the oil in physic nut (Jatropha curcas L.). Ciênc. Rural 40(8):1752-1758. |
|
|
Statsoft Inc. (2010). STATISTICA, versão 10. http://statistica.io/. |
|
|
Sunil N, Varaprasad KS, Sivaraj N, Kumar TS, Abrahan B, Prasad RBN (2008). Assessing Jatropha curcas L. germplasm in situ: a case study. Biomass Bioenergy 32(3):198-202. |
|
|
Trebbi D, Papazoglou EG, Saadaoui ES, Vischi M, Baldini VM, Stevanato P, Cettul E, Sanzone AP, Gualdi L, Fabbri A (2015). Assessment of genetic diversity in different accessions of Jatropha curcas. Ind. Crops Prod. 75:35-39. |
|
|
Tripathi A, Mishra DK, Shukla JK (2015). Genetic diversity and trait association between growth, yield and seed component of Jatropha curcas (L.) source collection from Indian sub-continent. J. Plant Breed. Crop Sci. 7(5):143-157. |
|
|
Vaknim Y, Ghanim M, Samra S, Dvash L, Hendelsman E, Eisikowitch D, Samocha Y (2011). Predicting Jatropha curcas seed-oil content, oil composition and protein content using near-infrared spectroscopy - A quick and non-destructive method. Ind. Crops Prod. 34(1):1029- 1034. |
|
|
Wani S, Garg KK, Chander G (2016). Water needs and productivity of Jatropha curcas in India: myths and facts. Biofuels Bioprod. Bioref. 10:240-254. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0