ABSTRACT
Acetic acid bacteria are known for their ability to oxidize ethanol to acetic acid. This study investigated to identify dominant strain acetic acid bacteria involving in fermented juice of mango and capable to produce Vinegar, total of 4 bacteria (CRSBAN-BVA1, CRSBAN-BVK1, CRSBAN-BVK2, CRSBAN-BVI1) bacteria strains were preselected ¶for the analyses. The strains were examined with biochemical, physiological and phenotypical methods such as Gram die, catalase and oxidase test, ethanol oxidation to acetic acid, and over to CO2 and H2O and also metabolism of carbohydrate was tested, for their affiliation to the genera of acetic acid bacteria. Subsequently, genotypic identification was conducted by sequencing the gene coding for 16S rRNA of one targeted strain and phylogenetic analysis was realized throughout 16S rRNA sequences. The results showed that one of the isolated strain (CRSBAN-BVA1) present 99. 90% of similarity in the sequence 16S rRNA region with Acetobacter tropicalis. It demonstrated that bacterial diversity in the mango vinegar is dominated by A. tropicalis. Therefore this strain is potentially useful for its utilization as a starter in vinegar production.
Key words: Fermented juice, acetic acid bacteria, 16S rRNA gene sequence, Acetobacter tropicalis.
Acetic acid bacteria (AAB) are important organisms in food and beverage industries etc. It is known that they adapt well to sugary and alcoholized fluid (Muramatsu et al., 2009). AAB are Gram negative, rod shape and obligate aerobic bacteria with the ability to oxidize ethanol to acetic acid (Moryadee et al., 2008; Maal et al., 2010). Belonging to the family of Acetobacteriaceae, AAB are classified in twelve genera: Acetobacter, Gluconobacter, Acidomonas, Gluconacetobacter, Asaia, Kozakia, Swaminathania, Saccharibacter, Neoasaia, Granulibacter, Tanticharoenia and Ameyamaea (Sengun et al., 2011). They are ubiquitous organisms that are well adapted to sugar and ethanol rich environments (Bartowsky et al., 2008). Vinegar, from the French vinaigre, meaning sour wine, can be made from almost any fermentable carbohydrate source, including apples, dates, grapes, pears, coconut, honey, mangos etc (Johnston and Gaas, 2006). Burkina Faso, as in other Sahelian countries, fruits production is dominated by mango. The production of fresh mangoes is estimated at around 337101 ton per year (CEFCOD, 2013). However, factors such as: lack of control over harvesting, packaging and storage standards, poor road infrastructure, poor commercialized on the local market and inadequacy of processing infrastructures, inflict enormous losses which handicap this sector. The resulting annual losses are estimated at about 30-40% of mango production (Ngamo et al., 2010; Vayssieres et al., 2013). However, mango is a substrat rich in fermentable substances sugars.
Thus, transforming them via biotechnological processes to obtain exotic products like vinegar by the local strain of AAB would be interressant. In Burkina like the other majority countries of West Africa, the most of vinegar consumed comes from the dilution of acetic acid of chemical origin because of lack of AAB strain. To raise this challenges two Acetobacter strains, Acetobacter tropicalis and Acetobacter pasteurianus were isolated Dolofrom mango fruit (Mangifera indica) in Senegal and (local beer obtained by fermenting cereal product) in Burkina Faso respectively (Ndoye et al., 2006).
This study aimed to isolate an AAB strain whose features make them applicable for biological vinegar production. Thus, we first isolated, identified and characterized AAB strains of mangos fruits. A recent classification of the AAB includes the genera of Acetobacter, Acidomonas, Ameyamaea, Asaia, Gluconacetobacter, Gluconobacter, Granulibacter, Kozakia, Neoasaia, Neokomagataea, Saccharibacter, Swaminathania and Tanticharoenia (Yamada and Yukphan, 2008; Mamlouk and Gullo, 2013).
AAB are generally found in nature because they can use a variety of substrates (Sharafi et al., 2010) and these bacteria have been isolated from alcoholic beverage, vinegar, fruits and fruit juice, flowers, honey, sugar cane, soil and water (Klawpiyapamornkun et al., 2015). Mango waste has a high carbohydrate and organic acid content that creates an acidic niche (Ouattara et al., 2018). Therefore, fermented juice of mango is a good source for isolation of AAB (Ouattara et al., 2018). The methods of identification based on the analysis of the phenotypical characteristics of the bacteria of non precise acetic acid and also very long do not have is not enough with the identification to the species. To optimize their use, it is necessary to determine their 16S rRNA gene sequences to understand their taxonomic positions.
Sampling
One kilogram of mango samples were collected from four sites in Burkina Faso (Bobo-Dioulasso, Banfora, Orodara and Ouagadougou) (Figure 1). Six different varieties (Amelie, Kent, Sauvage, Brooks, Lippens and Springfield) of mangos were collected. A total of 80 mangos samples of different varieties were collected in May-July 2016 and 2017. After collection, they were subsequently crushed aseptically and were stored for spontaneous fermentation at room temperature.
Screening of acetic acid bacteria (AAB)
Screening of potentially AAB was performed on GYEA modified medium. Prefermented mango were transferred in a GYEA enrichment medium containing of glucose 2% (w/v), yeast extract 1% (w/v), ethanol 2% (v/v) and acetic acid 1% (v/v). Samples were incubated under agitation (120 rpm) at room temperature (30°C) for one week (Mounir et al., 2016). A volume of 100 μl from different dilutions were inoculated in GYC solid medium (10% glucose, 1.0% yeast extract, 2.0% calcium carbonate, 1.5% agar, pH 6.8) supplemented with 100 mg l-1 of Cycloheximide and nystatine were to inhibit the growth of fungi and lactic acid bacteria, respectively (Sharafi et al., 2010; Kadere et al., 2008). This antibiotic was added to the culture medium from the stock solution after the medium had been sterilized. Plates dish were incubated at 30°C for 2-3 days under aerobic conditions. Only isolates which were able to produce clear halos around the colonies, caracteristic fundamental associates a colony to the group of acetic bacteria were further characterized (Cleenwerck and De Vos, 2008). Acetobacter and Gluconobacter were distinguished from each other on Carr medium in the presence of bromocresol green. Acetobacter turns the media color to yellow and then to green while Gluconobacter turns it into yellow.
Phenotypic characterization of acetic acid bacteria (AAB)
Biochemical and morphological identification tests were performed to confirm that the selected isolates belong to Acetobacter genera. Morphology of bacteria, including their shape, size, arrangement, Gram and motility, was characterized using cells grown on GYC at 30°C under aerobic conditions (Cleenwerck et al., 2002). Tests, such as catalase, oxidase, and growth in varying concentrations of ethanol and glucose, were employed according the method of Conventional biochemical. Other biochemical tests such as carbohydrate assimilation was performed on presumed Acetobacter strains.
DNA preparation
DNA extraction was performed according method by Ruiz et al., (2000) with the following modifications. Cells were grown overnight in 5 ml medium, centrifuged at 4000 g and washed twice with 5 ml of water. The pellet was suspended in 300 ml of 3% (w/v) SDS-TE buffer (10 mmol l-1 Tris-HCl, pH 7.5; mmol l-1EDTA) and incubated for 10 min at room temperature. After addition of 200 ml TE buffer, 500 ml of phenol-chloroform-isoamyl alcohol (25: 24: 1, v/v), the aqueous phase was separated by centrifugation for 10 min at 10 000 g. The DNA was precipitated with isopropanol and washed with 70% (v/v) ethanol. Finally, the DNA was resuspended in 20 ml TE buffer to a final concentration of 1150 ng/ml and stored at -20°C.
PCR amplification, sequencing, and phylogenetic analysis of 16S rRNA genes
DNA for 16S rRNA gene sequencing was extracted by the method of Wilson (2001) with minor modifications (Cleenwerck et al., 2002). The 16S rDNA genes of both community bacteria were amplified by PCR using the pair of 16S rDNA gene universal primers. Forward primer (5-AGAGTTTGATCCTGGCTCAG-3) and Reverse primer (5-ACGGCTACCTTGTTTACGACTT-3) which is targeted to bacterial 16S rDNA gene was used. The forward and reverse 16S rDNA gene universal primers generate a 1.5 kb fragment. The polymerase chain reaction (PCR) reaction was performed in 0.5 ml microcentrifuge tubes (Eppendorf, UK) with 25 ml of reaction mixture: 12.5 ml Ì Go Taq Green Ì‹ master mix (2.5 units Taq DNA polymerase, 1X Qiagen PCR buffer and 200 μM of each dNTP), 0.5 μl forward primer (10 μM), 0.3 μl reverse primer (10 μM) and 1.5 μl RNA template, and made up to 25 μl with 10.2 μl of nuclease-free sterile distilled water. The PCR protocol consisted of an initial denaturation step of 95°C for 5 min followed by 30 cycles of denaturation at 94°C for 1 min, primer annealing at 44°C for 30 s and elongation at 72°C for 2 min, final holding at 73 °C for 4 min. PCR reaction was performed in a 20 well block thermocycler (TECHGENE, UK). Sequence blast, alignment and phylogenetic trees were obtained from the website le BIBI https://umr5558-bibiserv.univ-lyon1.fr/lebibi/lebibi.cgi. The topology of the trees was evaluated using bootstrap method with 1000 replicates. Alignments of 16S rDNA sequences from the GenBank database were screened to select the most suitable primers to use in the detection and identiï¬cation of AAB.
Phenotypical identification of strains
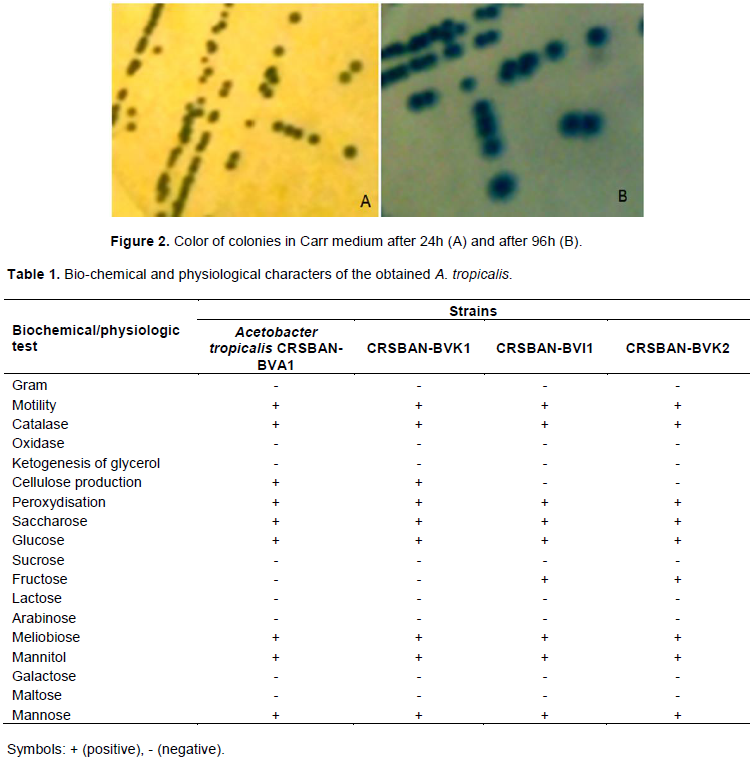
The examination of the primary screened such as macroscopic, microscopic and biochemical of isolated strain showed that this strain related to AAB group. Overoxidation in Carr medium resulted in conversion of blue color of medium to yellow after 24-48 h and then reconversion of yellow color to blue after 72-96 h (Figure 2). Also utilization of CaCO3 and creation of transparent zones around the colonies in Frateur medium confirmed that the isolated strain was AAB. All strains were able to produce acid from following sugars such as: glucose, mannose, melibiose and mannitol and were unable to produce acid from arabinose, galactose, fructose, lactose, maltose, sucrose and saccharose. The preliminary identification on the basis of biochemical and physiologique tests (Table 1) brought about the possibility of having the Acetobacter. Hence the 16s rRNA technique was further employed to confirm the isolate.
Identification of A. tropicalis
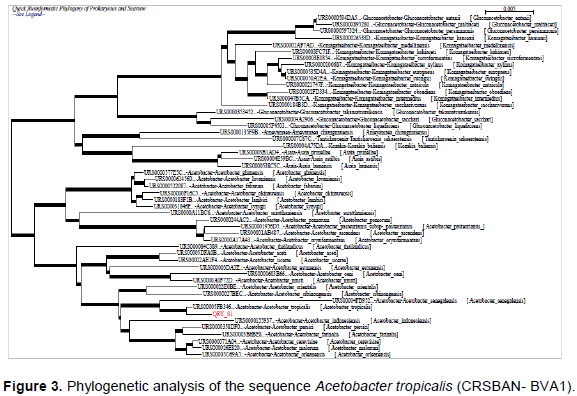
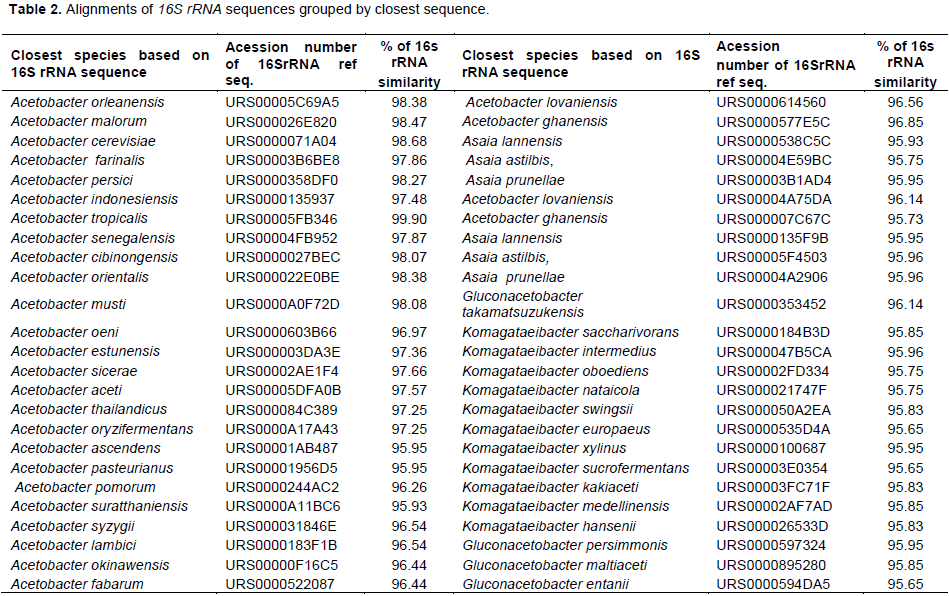
The phylogenetic affiliation of strain was based on 16S rRNA gene sequence analysis, where it was shown to belong to the genus Acetobacter. The phylogenetic analysis of the strains (Figure 3) was carried out using leBiBi software to determine similarity and close relationship of isolate. The phylogenetic tree analysis revealed that sequence was closely related to Acetobacter species. Figure 3 showed that strain A. tropicalis (CRSBAN-VBA1) belonged to the stable subcluster containing A. orleanensis, A. malorum, A. cerevisiae, A. farinalis, A. persici, A. indonesiensis, A. tropicalis, A. senegalensis, A. cibinongensis, A. orientalis, A. musti, A. oeni, A. estunensis, A. sicerae, A. aceti, A. thailandicus, A. oryzifermentans, A. ascendens, A. pasteurianus, A. pomorum A. suratthaniensis, A. syzygii, A. lambici, A. okinawensis A. fabarum, A. lovaniensis and A. ghanensis. The 16S rRNA gene sequence similarities obtained by pairwise alignment with the Bio Numerics 4.5 software package between strain A. tropicalis (CRSBAN-BVA1) and the type strains or another strains of recognized Acetobacter species were represented in Table 2.
Acetic acid bacteria are characterized by the ability to oxidize alcohols or sugars incompletely, and a common feature to most of strains their capacity to oxidize ethanol to acetic acid. Although strains of AAB that are generally isolated with GYC plates showed distinct clear zones (Trcek, 2005). The methods based on the presence of a clear zone was not completely believable because other strains, such as some lactic acid bacteria, could also form distinct clear zones (Trcek, 2005). The result of biochemical tests showed that the isolated strains from fermented juice of mangos belonged to genus of Acetobacter or Gluconacetobacter. In addition to their ability to oxidise ethanol, Acetobacter and Gluconacetobacter species can further oxidise acetic acid to CO2 and H2O, generating the so-called acetate overoxidation, that is carried out by the tricarboxylic acid cycle (TCA) when there is a high level of dissolved oxygen and no ethanol in the medium. According to Sakurai et al., (2013), strains of Gluconobacter are not able to overoxidise because of non-functional α-ketoglutarate dehydrogenase and succinate dehydrogenase of tricarboxylic acid cycle; they can only oxidize ethanol to acetic acid (Du Toit and Pretorius, 2002). Hence the presence of the ethanol in the medium represses the activity of TCA enzyme cycles in Acetobacter genus. The results of acid production with different sugars showed also that strains produced acid with some sugars and did not have the capacity to produce acid with other sugars. These results are slightly comparable to those found by Lisdiyanti et al. (2000) and kadere et al. (2008). According to Lisdiyanti et al., (2000) this slight difference is a variability between strains of Acetobater genus.
On the Carr medium, the production of the acid in the medium by strain can be seen in form of clearing of opacity of medium or the colour change of bromocresol green confirms that the isolate is Acetobacter species. Sharafi et al., (2010); Mounir et al., (2016) had reported that color change of the indicator bromocresol green in the medium from green to yellow confirm that the isolate is Acetobacter. The production of acid acetic of this strains was previously determined by Ouattara et al., (2018), where it was shown that these strains had the capacity to produce a high concentration of acetic acid and the highest was found with strain CRSBAN-BVA1. Molecular characterization will confirm our results. Molecular techniques have been employed by PCR-amplified fragment of the gene coding for 16S rRNA to confirm that the isolate is Acetobacter.
The fast molecular detection was proven to be efficient and accurate According 16S rRNA sequencing and phylogenetic tree analysis, the isolate was further proven to be A. tropicalis. The comparison of 16S rRNA gene sequence of the strains with the total nucleotide collection in the LEBIBI nucleotide database was used to assign the bacterial name with ≥ 99% similarity. The greatest similitude (99.90%) was obtained with the species A. tropicalis and another similitude which various between 95.75 to 98.68% with other species such as Acetobacter, Asaia, Gluconacetobacter and Komagataeibacter were represented in Table 2. A. tropicalis was identified as the only predominant group. The phylogenetic tree reflects the results obtained in Table 2. Lisdiyanti et al., (2000) in their study on the diversity of AAB in Indonesia, Thailand and the Philippines, have isolated A. tropicalis from fermented foods (palm wine and rice wine), fruits (lime, orange, guava, coconut), and coconut juice whose similarity is in a range of 96.5 to 98.9% between the type strain of A. tropicalis and the type strains of other Acetobacter species. Others researchers as Kounatidis et al., (2009) have isolated A. tropicalis with in similitude 99.7% from fermented wine. In Senegal A. tropicalis with in similitude 99.3% was isolated from mango fruit (Mangifera indica) (Ndoye et al., 2007). These similarities are slightly lower than our which was 99.90%. Hence the identity of the isolates were confirmed by the 16s rRNA method, as A. tropicalis.
In this study, AAB were isolated and identified from fermented juice of mangos sing of 16S rRNA gene sequence has allowed differentiation between species and could represent a tool for a rapid and low cost effective preliminary profiling of Acetic Acid Bacteria genera. The molecular technique can also be useful to highlight the phylogenetically closely related species. This first knowledge of the acetic acid bacteria will serve as a guide in selecting starter for the production of vinegar in Burkina Faso.
The authors have not declared any conflict of interests.
The authors need to thank Laboratory Microbial Biotechnology then Laboratory of Food Technology (Department of Biochemistry and Microbiology, University of Joseph KI-ZERBO), Ministry for the Woman, national Solidarity, the Family and the humane Action for their help and Laboratory of Genoscreen for genotyping. We also thank the International Sciences Programme (ISP-Sweden).
REFERENCES
|
Bartowsky EJ, Henschke PA (2008). Acetic acid bacteria spoilage of bottled red wine-Areview. International Journal Food of Microbiology 125(1):60-70.
Crossref
|
|
|
|
Centre d'Etude, de Formation et de Conseil en Développement (CEFCOD) (2013).Situation de référence des principales filières agricoles au Burkina Faso. Rapport version finale, Ministère de l'Agriculture et de la Sécurité Alimentaire 74 p.
|
|
|
|
|
Cleenwerck I, De Vos P (2008). Polyphasic taxonomy of acetic acid bacteria: An overview of the currently applied methodology. International Journal of Food Microbiology 125(1):2-14.
Crossref
|
|
|
|
|
Cleenwerck I, Vandemeulebroecke K, Janssens D, Swings J (2002). Re-examination of the genus Acetobacter, with descriptions of Acetobacter cerevisiae sp. nov., and Acetobacter malorum sp. nov. International Journal of Systematic and Evolutionary Microbiology 52(5):1551-1558.
Crossref
|
|
|
|
|
Du Toit WJ, Pretorius IS (2002). The occurrence, control and 12 esoteric effect of acetic acid bacteria in winemaking. Annals of Microbiology 52(2):155-179.
|
|
|
|
|
Johnston CS, Gaas CA (2006). Vinegar: medicinal uses and antiglycemic effect. Medscape General Medicine 8(2):61. PMID:16926800.
|
|
|
|
|
Kadere TT, Miyamoto T, Oniang R K, Kutima PM Njoroge SM (2008). African Journal of Biotechnology 7(16):2963-2971.
|
|
|
|
|
Klawpiyapamornkun T, Bovonsombut S, Bovonsombut S (2015). Isolation and Characterization of Acetic acid Bacteria from Fruits and Fermented fruit juices for Vinegar Production. Food and Applied Bioscience Journal 3(1):30-38.
|
|
|
|
|
Kounatidis I , Crotti E, Sapountzis P, Sacchi L, Rizzi A, Chouaia B, Bandi C, Alma A, Daffonchio D, Mavragani TP, Bourtzis K (2009). Acetobacter tropicalis Is a Major Symbiont of the Olive Fruit Fly (Bactrocera oleae). Applied and Environmental Microbiology 75(10):3281-3288.
Crossref
|
|
|
|
|
Lisdiyanti P, Kawasaki H, Seki T, Yamada Y, Uchimura T, Komagata K (2000). Systematic study of the genus Acetobacter with descriptions of Acetobacter Indonesiensis sp. nov., Acetobacter tropicalis sp. nov., Acetobacter orleanensis (Henneberg 1906) comb. nov., Acetobacter lovaniensis (Frateur 1950) comb. nov., and Acetobacter estunensis (Carr 1958) comb. nov. Journal of General and Applied Microbiology 46(3):147-165. PMID:12483588.
Crossref
|
|
|
|
|
Maal KB, Shafiei R, Kabir N (2010). Production of Apricot Vinegar Using an Isolated Acetobacter Strain from Iranian Apricot. International Journal of Food nutrition and food Engineering 4(11):177-180.
|
|
|
|
|
Mamlouk D, Gullo M (2013). Acetic Acid Bacteria: Physiology and Carbon Sources Oxidation. Indian Journal of Microbiology 53(4): 377-384.
Crossref
|
|
|
|
|
Moryadee A, Pathom-Aree W (2008). Isolation of Thermotolerant Acetic Acid Bacteria From Fruits for Vinegar Production. Journal of Microbiology 3(3): 209-212.
Crossref
|
|
|
|
|
Mounir M, Shafiei R, Zarmehrkhorshid R, Hamouda A, Ismaili AM, Thonart P (2016). Simultaneous production of acetic and gluconic acids by a thermotolerant Acetobacter strain during acetous fermentation in a bioreactor. Journal of Bioscience and Bioengineering 121(2):166-171.
Crossref
|
|
|
|
|
Muramatsu Y, Yukphan P, Takahashi M, Kaneyasu M, Malimas T, Potacharoen W, Yamada Y, Nakagawa Y, Tanticharoen M, Suzuki K (2009). 16S rRNA gene sequences analysis of acetic acid bacteria isolated from Thailand. Microbiology and Culture Collections 25(1): 13-20. PMID:19590153.
|
|
|
|
|
Ndoye B, Weekers F, Diawara B, Guiro AT, Thonart P (2007). Survival and preservation after freeze-drying process of thermoresistant acetic acid bacteria (TAAB) isolated From tropical products of Subsaharan. African Journal Food Engineering 79(4):1374-1382.
Crossref
|
|
|
|
|
Ndoye B, Lebecque S, Dubois-Dauphin R, Tounkara L, Guiro TA, Kere C, Diawara B,Thonart P (2006). Thermoresistant properties of acetic acid bacteria isolated from tropical products of Sub-Saharan Africa and destined to industrial vinegar. Enzyme and Microbial Technology 39(4):916-923.
Crossref
|
|
|
|
|
Ngamo TL, Ladang D, Vayssieres JF, Lyannaz JP (2010). Diversité des espèces de mouches des fruits (Diptera :Tephritidae) dans un verger mixte dans la localité de Malang (Ngaoundéré, Cameroun). International Journal of Biological and Chemical Sciences 4(5):1425-1434.
Crossref
|
|
|
|
|
Ouattara A, Somda KM, Ouattara ATC, Traore SA, Ouattara SA (2018). Production of acetic acid by acetic acid bacteria using mango juice in Burkina Faso). International Journal Biological and Chemical Science 12(5): 2309-2317.
Crossref
|
|
|
|
|
Ruiz A, Poblet M, Mas A, Guillamon JM (2000). Identification of acetic acid bacteria by RFLP of PCR-amplified 16S rDNA and 16S-23S rDNA intergenic spacer. International Journal of Systematic and Evolutionary Microbiology 50(6):1981-1987.
Crossref
|
|
|
|
|
Sakurai K, Yamazaki S, Ishii M, Igarashi Y, Arai H (2013). Role of the glyoxylate pathway in acetic acid production by Acetobacter aceti. Journal of Bioscience and Bioengineering 115(1):32-6.
Crossref
|
|
|
|
|
Sharafi SM, Rasooli I, Beheshti-Maal K (2010). Isolation, characterization and optimization of indigenous acetic acid bacteria and evaluation of their preservation methods. Iranian journal of Microbiology 2(1):38-45. PMID: 22347549.
|
|
|
|
|
Sengun IY, Karabiyikli S (2011). Importance of acetic acid bacteria in food. Industry Food Control 22(5):647-656.
Crossref
|
|
|
|
|
Trcek J (2005) .Quick identification of acetic acid bacteria based on nucleotide sequences of the 16S-23S rDNA internal transcribed spacer region and of the PQQ-de¬pendent alcohol dehydrogenase gene. Systematic and Applied Microbiology 28(8):735-745.
Crossref
|
|
|
|
|
Vayssieres JF, Sinzogan A, Adandonon A, Van Mele Paul, Korie S (2013). Ovipositional behaviour of two mango fruit fly species (Diptera Tephritidae) in relation to Oecophylla cues (Hymenoptera Formicidae) as compared to natural conditions without ant cues. International Journal of Biological and Chemical Sciences 7(2):447- 456.
Crossref
|
|
|
|
|
Wilson K (2001). Preparation of genomic DNA from bacteria. In Current Protocols in Molecular Biology 56(1):2. 4.1-2.4.5.
Crossref
|
|
|
|
|
Yamada Y, Yukphan P (2008). Genera and species in acetic acid bacteria. International Journal Food of Microbiology 125(1):15-24.
Crossref
|
|