ABSTRACT
An experimental protocol originally designed to isolate plant DNA was modified to obtain high quality total RNA from organs of adult coconut palms collected in situ. With this protocol, high quality RNA was extracted from leaves, inflorescences, primary and secondary roots, zygotic embryos and solid endosperm, with no carbohydrate or protein contamination. Reverse transcription-polymerase chain reaction (RT-PCR) amplification of a 470 bp cDNA, corresponding to a highly conserved domain of the eukaryotic mitogen-activated protein kinases, demonstrated the integrity of the RNA samples. Isolation of intact RNA from coconut palms growing under wild conditions facilitates the study of gene regulation ex vivo.
Key words: Coconut palms, RNA extraction, secondary metabolites.
Abbreviation:
CTAB, Cethyl-trimethyl-ammonium bromide; PVP, polyvinylpyrrolidone; FL, flag leaf; IF, immature inflorescence; SR, secondary root; ZE, zygotic embryo; SE, solid endosperm, MAPK, mitogen-activated protein kinase.
Mangrove and coconut palms are key elements that protect the coastal zones and tropical wetlands against the environmental impacts. Cultivation of coconut represents a major income for the people in the tropical and subtropical zones of the world (Mathew, 1986; Hyman, 1990); however, plantations are continuously threatened by different pests and diseases that reduce crop productivity (Zizumbo-Villarreal et al., 2006; Magalhães et al., 2008). Breeding of the coconut palms to generate stress-resistant varieties by means of biotechnological methods has been delayed due to their recalcitrance to cultivation in vitro (McCown, 2000). Also, their organs contain secondary metabolites that potentially interfere with the RNA isolation. The leaves are rich in lignin and their surfaces have a dense coat of epicuticular waxes (Escalante-Erosa et al., 2007); the inflorescences contain high amounts of lignin fibers, which have been found to reinforce epoxy composite materials (Sapuan et al., 2005); the embryos are specialized in the accumulation of lipids and carbohydrates (Sugimura and Murakami, 1990; López-Villalobos et al., 2001); and the roots possess a complex structure and composition that allow the palm to struggle with the high saline environment found in the coast soils (Nainanayake et al., 2000). High polyphenol contents in adult tissues become evident because activated charcoal must be added to the culture media when adult tissues are used as explants for somatic embryogenesis in vitro (Gupta et al., 1984; Chan et al., 1998).
The isolation of coconut ribonucleic acid (RNA) from soft tissues, young seedlings and calli cultivated can be performed by the use of standard protocols that are based on the use of guanidine thiocyanate (Chomczynski and Sacchi, 1987) or cetyltrimethylammonium bromide (CTAB) detergent (Xiao et al., 2012; Gao et al., 2014; Liang et al., 2014; Yuan et al., 2015). Lizama et al. (2007) analyzed the molecular regulation of disease responses by comparing transcript populations isolated from chitosan-elicited in vitro coconut calli. In a different study, Pérez-Núñez et al. (2009) quantified transcript levels of a gene encoding a receptor-like kinase during the development of coconut embryogenic calli. Rajesh et al. (2015) isolated total RNA from embryogenic calli and characterized the global transcriptome of coconut palm (Cocos nucifera L.) during somatic embryogenesis. In this study, the isolation of high quality RNA was performed by the use of Trizol® reagent (Invitrogen). Other commercial protocols the like RNeasyTM Plant Kit (Qiagen) or the Plant Total RNA Miniprep Purification KitTM (GMbiolab Co., Ltd.) have also been used to isolate RNA from embryonic tissues (Bandupriya et al., 2014) or seedling leaves (Huang et al., 2013), to determine the expression of a coconut homeotic gene in zygotic and somatic embryos and during germination, and to analyze the chloroplast genome of the coconut palm, respectively.
An effort had been made in the laboratory to isolate total RNA from different organs of adult palms growing in the coasts. However, neither of the protocols reported above nor other protocols designed to isolate RNA from woody or secondary metabolite-rich plants yielded RNA from several organs of adult palms (Jaakola et al., 2001; Valenzuela-Avendaño et al., 2005), with the minimum quality even visualized in agarose gels. Thus, a specific protocol for the isolation of high quality total RNA from adult coconut palms was established, by complementing reported protocols with modifications devoted to eliminate interfering contaminants during the isolation of RNA. In the present work, the isolation of high quality total RNA of different organs from adult coconut palms collected in situ was reported, by the modification of a CTAB method designed to extract RNA from plant tissues with high phenolic compounds, polysaccharides and elevated levels of RNases (Jaakola et al., 2001). The purity and integrity of the RNA samples was evaluated spectrophotometrically and by electrophoretic fractionation in agarose gels. The integrity of the isolated RNAs samples was confirmed by the successful reverse transcription-polymerase chain reaction (RT-PCR) amplification of a complementary deoxyribonucleic acid (cDNA) fragment corresponding to a coconut mitogen-activated protein kinase (MAPK) transcript.
Plant
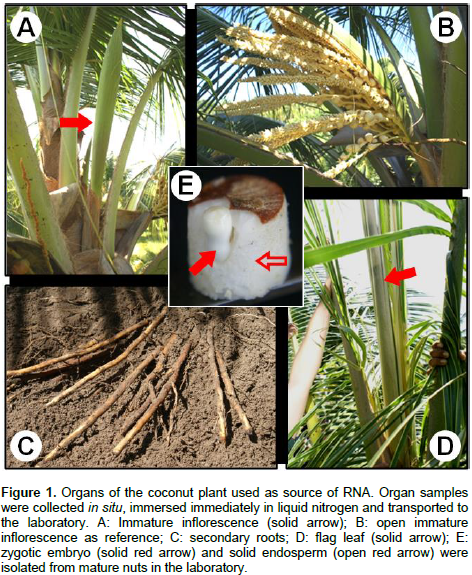
The coconut immature inflorescences (physiological state (PS) = -4, numbered regressively from the last open inflorescence), the pine group of the last emitted leaf (flag leaf), and the meristematic zone of primary and secondary roots were collected from adult palms of the “Atlantic Tall” variety, cultivated in the San Crisanto town in the north coast of the Yucatan Peninsula, Mexico (21°, 21´ 00.74´´ N; 89°, 11´32.49´´ O). All tissues were immediately frozen in liquid nitrogen and transported to the laboratory, and then they were stored at -80°C until processed. Mature nuts were collected in situ and transported to the laboratory to dissect the zygotic embryos and the solid endosperm by the method reported by Chan et al. (1998).
RNA extraction
RNA extraction was attempted following different methodologies. Trizol® reagent, TRI Reagent® and Concert® were used accordingly to the manufacturer’s instructions. The CTAB method reported by Jaakola et al. (2001) and the method reported by Valenzuela-Avendaño et al. (2005) were followed as reported. The CTAB modified method presented here was performed as follows. 250 mg of each coconut tissue were ground to powder in liquid nitrogen. Then 1 mL of the CTAB solution (2% cethyl-trimethyl-ammonium bromide; 2 M NaCl; 20 mM EDTA, pH 8; 100 mM Tris-HCl, pH 8, with freshly added β-mercaptoethanol and polyvinylpyrrolidone-40 (PVP-40) to 2% final concentration each) was added and the solution was thoroughly homogenized. The crude extracts were halved into two 1.5 mL Eppendorf tubes and then they were mixed vigorously for 10 min at room temperature. The samples were centrifuged at 12,000 ×g for 15 min at room temperature. The supernatants were transferred to new tubes and then they were extracted twice with one volume of a chloroform: isoamyl alcohol solution (49:1) and centrifuged at 12,000 ×g for 10 min at room temperature.
The last supernatants were transferred to clean tubes and mixed perfectly with 0.5 volumes of ice-cold isopropanol; then, 0.5 volumes were added of the saline solution (0.8 M sodium citrate/ 3 M NaCl) and the tubes were gently mixed for 10 min. The solution was centrifuged at 12000 ×g for 10 min at 4°C. The pellets were washed twice with 75% ethanol and centrifuged at 12000 ×g for 10 min at 4°C and they were air-dried for 10 min at room temperature. The pellets were dissolved in 100 μL of H2O-DEPC, and then they were mixed with 264 µL of ice-cold 4 M LiCl (2.85 M final concentration) and stored for 1 h on ice. After a centrifugation at 12000 xg for 10 min at 4°C, the RNA pellets were washed twice with 75% ethanol and they were air-dried for 10 min at room temperature. The pellets were dissolved in 30 μL of H2O-DEPC free (SIGMA). The integrity and purity of the RNA preparations were assessed by electrophoretic fractionation in agarose gels, and by measuring the optical density at 230, 260 and 280 nm, respectively.
Reverse transcription-polymerase chain reaction (RT-PCR)
To evaluate the functionality of the RNA preparations, 0.8 μg of total RNA isolated from each organ sample were used as template for the RT-PCR amplification of a 470 bp cDNA fragment, corresponding to the conserved domain of mitogen-activated protein kinases. The Superscript One-Step RT-PCR kit with Platinum Taq polymerase® (Invitrogen) was used as recommended by the manufacturer, using the degenerate primers 5’-GGNGCYTACGGHATYGTTTGYTCK-3’ (forward) and 5’- GGNGCYTACGGHATYGTTTGYTCK -3’ (reverse), under the following cycling conditions: one cycle at 42°C, 2 min; 48°C, 30 min; 94°C, 2 min; then, forty cycles at 94°C, 15 s; 50°C, 30 s; 72°C, 1 min, followed by a final extension step at 72°C for 10 min. 3 μL aliquots of each RT-PCR product were fractionated by agarose gel electrophoresis, and then the gel was stained with 1 μg ml-1 ethidium bromide and visualized under UV light.
The isolation of high quality RNA is an essential step to carry out molecular studies in plants; however, the extraction of RNA is compromised in plant tissues rich in secondary metabolites and complex carbohydrates (Jaakola et al., 2001). This is the case of the coconut palm; indeed, the breeding of elite coconut varieties has been hampered because of several adverse factors inherent to the adult palm, including its size and long life cycle. In addition, the coconut palm is a perennial monocot woody plant with high recalcitrance to in vitro cultivation (McCown, 2000). Furthermore, the difficulty of isolating biological molecules from coconut can be evidenced by the existence of extremely few reports in this area. In the laboratory, investigation of the regulation of the molecular responses of coconut cells to the presence of pathogenic signals in the environment was done. While, the use of coconut tissues growing in vitro has been suggested as an alternative to study the gene regulation (Chakraborty et al., 2009; Lizama et al., 2007), the analysis of gene function must be done in whole palms, requiring the extraction of RNA from the organs of interest Adult organs of coconut palms growing in the field was collected (Figure 1) and tested different commercial methods to isolate total RNA; however, their use yielded RNA samples that could not be visualized [Trizol® (Invitrogen), TRI Reagent® (SIGMA-ALDRICH)] or appeared as smears in agarose gels [Concert® (Invitrogen)] (Figure 2, TZ, TR and CN, respectively).
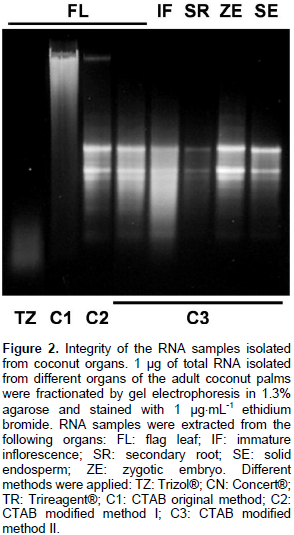
Conversely, the use of CTAB method as described by Jaakola et al. (2001) yielded only DNA (Figure 2, lane C1). It has been proposed that the CTAB effect during the extraction of nucleic acids from plants resides in its capacity to bind anionic polymers (that is glucuronoarabinoxylans) (Kiefer et al., 2000). The authors decided to modify the CTAB protocol to increase its capacity to eliminate phenolic compounds and complex carbohydrates by the addition of polyvinylpyrrolidone and a further precipitation step in the presence of a high concentration of salts. The addition of PVP during the extract preparation, and the extraction of the cleared crude extract with a mixture of chloroform: isoamyl alcohol (49:1) followed by the precipitation of carbohydrates from the aqueous phase with a saline solution (0.8 M sodium citrate/3 M NaCl), and a final precipitation step with isopropanol were determinant to precipitate integral RNA from all samples (Figure 2, lane C2). The soluble nature of PVP could extend its capacity to form complex with phenolic compounds, preventing their union and the further oxidation of the RNA samples (Bekesiova et al., 1999). In this protocol, it was not necessary to heat the CTAB extracts at 65°C. It has been reported that precipitation of aqueous extracts with high concentrations of salts (1 M NaCl) favours the elimination of polysaccharides from genomic DNA (Fang et al., 1992) and from RNA (Valenzuela et al., 2005).
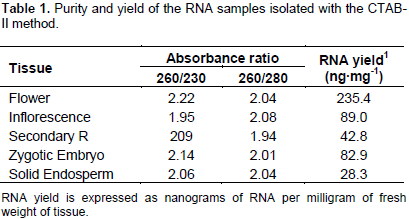
However, with these concentrations of salt, the spectrophotometric measurements gave 260/230 absorbance ratios ≤ 1.5 (data not shown). The increment of the NaCl concentration to 3 M (0.8 M sodium citrate/3 M NaCl) during the precipitation step produced RNA samples with 260/230 absorbance ratios @ 2. The contaminating DNA was eliminated from the salt-cleared RNA samples by a further precipitation with ice cold LiCl (2.85 M final) (Figure 2, lanes C3). Also, it was found that incubation of the LiCl-RNA mix at -20°C was not necessary because incubation on ice for 1 h produced high RNA yields. All preparations obtained with the new modifications yielded RNA with high integrity, as estimated by the band integrity of the major ribosomal RNAs (Figure 2). The yield and purity of the RNA samples were evaluated by spectrophotometric absorbance at 230, 260 and 330 nm. Table 1 shows that both 260:230 and 260:280 absorbance ratios were around the value of 2.0, implicating no significant contamination of the RNA samples with carbohydrates and proteins, respectively.
The modified CTAB method gave good RNA yields, especially from the palm flag leaf (235.4 ng mg-1). The smaller yield was obtained from solid endosperm secondary roots (28.3 ng mg-1). The quality of the RNA populations isolated from in situ collected adult organs was assessed by their capacity to function as template for reverse transcription in vitro. As can be seen in Figure 3, a single band was obtained by reverse transcription coupled with the polymerase chain reaction, using a pair of DNA oligonucleotides flanking a470 bp fragment of the ribonucleotide sequence encoding part of catalytic domain of the universally-conserved eukaryotic mitogen-activated protein kinases (MAPK). This result confirmed that the improved CTAB method yielded RNA extracts from adult palms cultivated in their natural ecosystems, with the quality required to perform molecular biology experiments required in modern biotechnological breeding programs.
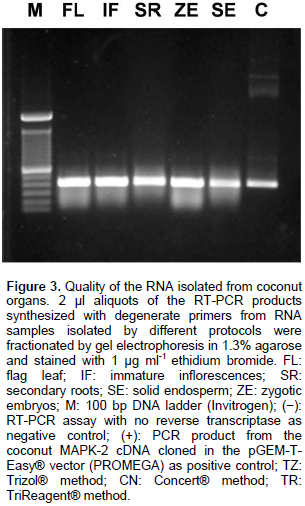
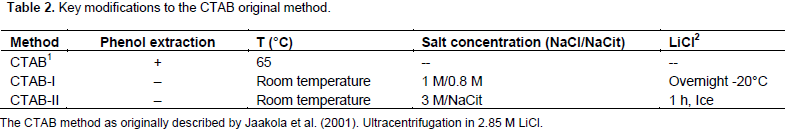
It is interesting to note that although the RNA samples obtained with the Concert® protocol served as templates to obtaining an RT-PCR amplicon of the expected size (Figure 3, lane CN), they appeared smeared after electrophoretic fractionation in agarose gels (Figure 2, lane CN), and their absorbance ratios 260/230 nm were below 1.0 units (data not shown). These results suggested contamination of the Concert® RNA sample with carbohydrates, which did not interfere with RT-PCR experiments, but affected electrophoretic mobility. Recently, few protocols to isolate RNA populations from soft tissues of coconut palms have been reported (Xiao et al., 2012; Bandupriya et al., 2014; Gao et al., 2014; Liang et al., 2014; Rajesh et al., 2015; Yuan et al., 2015), however, they have not been applied to hard tissues of coconut palm, like inflorescences or roots. The protocol presented here utilizes economic and easy-to find chemical ingredients; it could be applied to small amounts of tissue, reducing the cost of transportation and the amount of liquid nitrogen required to preserve samples collected in the field. It does not employ toxic chemicals (guanidine isothyocianate) or organic solvents (phenol) (Table 2), and it could be applied, with simple modifications, to the isolation of both DNA and RNA from the same coconut tissue.
The authors have not declared any conflict of interest.
The authors wish to thank the valuable support from coconut producers of San Crisanto Town in Yucatan, Mexico.
REFERENCES
|
Bandupriya HD, Gibbings JG, Dunwell JM (2014). Overexpression of coconut AINTEGUMENTA-like gene, CnANT, promotes in vitro regeneration in transgenic Arabidopsis. Plant Cell Tiss. Org. Cult.
Crossref
|
|
|
|
116:67-79.
|
|
|
|
|
Bekesiova I, Nap JP, Mlynarova L (1999). Isolation of High Quality DNA and RNA from Leaves of the Carnivorous Plant Drosera rotundifolia. Plant Mol. Biol. Rep. 17:269-277.
Crossref
|
|
|
|
|
Chakraborty M, Karun A, Mitra A (2009). Accumulation of phenylpropanoid derivatives in chitosan-induced cell suspension culture of Cocos nucifera. J. Plant Physiol. 166:63-71.
Crossref
|
|
|
|
|
Chan JL, Sáenz L, Talavera C, Hornung R, Robert M, Oropeza C (1998). Regeneration of Coconut (Cocos nucifera L.) from plumule explants through somatic embryogenesis. Plant Cell Rep. 17:515-521.
Crossref
|
|
|
|
|
Chomczynski P, Sacchi N (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156–159.
Crossref
|
|
|
|
|
Escalante-Erosa F, Arvízu-Méndez GE, Pe-a-Rodríguez LM (2007).The skimmiwallinols-minor components of the epicuticular wax of Cocos nucifera. Phytochem. Anal. 18:188-192.
Crossref
|
|
|
|
|
Fang G, Hammar S, Grumet R (1992). A quick and inexpensive method for removing polysaccharides from plant genomic DNA. Biotechniques13: 52-54.
|
|
|
|
|
Gao L, Sun R, Liang Y, Zhang M, Zheng Y, Li D (2014). Cloning and functional expression of a cDNA encoding stearoyl-ACP Δ9-desaturase from the endosperm of coconut (Cocos nucifera L.). Gene 549:70-76.
Crossref
|
|
|
|
|
Gupta PK, KendurkarSV, KulkarniVM, ShirgurkarMV, Mascarenhas, AF (1984).Somatic embryogenesis and plants from zygotics embryos of coconut (Cocos nucifera L.) in vitro. Plant Cell Rep. 3:222-225.
Crossref
|
|
|
|
|
Hyman EL (1990). The choice of technology and scale in coconut processing in the Philippines. Oléagineux 45:279-294.
|
|
|
|
|
Huang YY, Matzke AJ., Matzke M (2013). Complete sequence and comparative analysis of the chloroplast genome of coconut palm (Cocos nucifera). PLoS One 8:e74736.
Crossref
|
|
|
|
|
Islas-Flores I, Oropeza C, Hernández-Sotomayor SMT (1998).Protein phosphorylation during coconut zygotic embryo development. Plant Physiol. 118:257-263.
Crossref
|
|
|
|
|
Jaakola L, Pirttilä AM, Halonen M, Hohtola A (2001). Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol. Biotechnol. 19: 201-203.
Crossref
|
|
|
|
|
Kiefer E, Heller W, Ernst D (2000). A simple and efficient protocol for isolation of functional RNA from plant tissues rich in secondary metabolites. Plant Mol. Biol. Rep. 18:33-39.
Crossref
|
|
|
|
|
Liang Y, Yuan Y, Liu T, Mao W, Zheng Y, Li D (2014). Identification and computational annotation of genes differentially expressed in pulp development of Cocos nucifera L. by suppression subtractive hybridization. BMC Plant Biol. 14:1.
Crossref
|
|
|
|
|
Lizama-Uc G, Estrada-Mota IA, Caamal-Chan MG, Souza-Perera RA, Oropeza-Salin C, Islas-Flores I, Zú-iga-Aguilar JJ (2007). Chitosan activates a MAP-kinase pathway and modifies abundance of defense-related transcripts in calli of Cocos nucifera L. Physiol. Mol. Plant Physiol. 70:130-141.
Crossref
|
|
|
|
|
López-Villalobos A, Dodds PF, Hornung R (2001). Changes in fatty acid composition during development of tissues of coconut (Cocos nucifera L.) embryos in the intact nut and in vitro. J. Exp. Bot. 52:933-942.
Crossref
|
|
|
|
|
Magalhães JAS, De Moraes Neto AHA, Miguens FC (2008). Nematodes of Rhynchophorus palmarum, L. (Coleoptera: Curculionidae), vector of the Red Ring disease in coconut plantations from the north of the Rio de Janeiro State. Parasitol. Res. 102:1281-1287.
Crossref
|
|
|
|
|
Mathew A (1986). Coconut Economy of Kerala. Soc. Sci. 14:59-70.
Crossref
|
|
|
|
|
McCown BH (2000). Special symposium: In vitro plant recalcitrance of woody and herbaceous perennial plants: Dealing with genetic predeterminism. In Vitro Cell Dev-Pl 36:149-154.
|
|
|
|
|
Nainanayake NPAD, Bandara DC, Nissanka SP (2000). Root shoot relationships: an effective indicator of soil compaction and water stress for coconut (Cocos nucifera L.) seedlings. J. Trop. Agric. Res. 12:151-162.
|
|
|
|
|
Pérez-Nú-ez MT, Souza R, Sáenz L, Chan JL, Zú-iga-Aguilar JJ, Oropeza C (2009). Detection of a SERK -like gene in coconut and analysis of its expression during the formation of embryogenic callus and somatic embryos. Plant Cell Rep. 28:11-19.
Crossref
|
|
|
|
|
Rajesh MK, Fayas TP, Naganeeswaran S, Rachana KE, Bhavyashree U, Sajini KK, Karun A (2015). De novo assembly and characterization of global transcriptome of coconut palm (Cocos nucifera L.) embryogenic calli using Illumina paired-end sequencing. Protoplasma pp.1-16.
|
|
|
|
|
Sapuan SM, Zan MNM, Zainudin ES, Arora PR (2005). Tensile and flexural strengths of coconut spathe-fibre reinforced epoxy composites. J. Trop. Agric. 43:63-65.
|
|
|
|
|
Sugimura Y, Murakami T (1990). Structure and function of the haustorium in germinating coconut palm seed. Jpn Agricult. Res. Quart. 24:1-14.
|
|
|
|
|
Xiao Y, Yang Y, Cao H, Fan H, Ma Z, Lei X, Mason AS, Xia Z, Huang X (2012). Efficient isolation of high quality RNA from tropical palms for RNA-seq analysis. Plant Omics 5:584.
|
|
|
|
|
Yuan Y, Liang Y, Gao L, Sun R, Zheng Y, Li D (2015). Functional heterologous expression of a lysophosphatidic acid acyltransferase from coconut (Cocos nucifera L.) endosperm in Saccharomyces cerevisiae and Nicotiana tabacum. Sci Hort. 192:224-230.
Crossref
|
|
|
|
|
Valenzuela-Avenda-o JP, Estrada-Mota IA, Lizama-Uc G, Souza-Perera RA, Valenzuela-Soto EM, Zú-iga Aguilar JJ (2005). Use of a simple method to isolate intact RNA from partially hydrated Selaginella lepidophylla plants. Plant Mol. Biol. Rep. 23:199a-199g.
Crossref
|
|
|
|
|
Zizumbo-Villarreal D, Ruiz-Rodriguez M, Harries H, Colunga-García Marín P (2006). Population Genetics, Lethal Yellowing Disease, and Relationships among Mexican and Imported Coconut Ecotypes. Crop Sci. 46:2509-2516.
Crossref
|
|