The main objective of this work was to identify novel lipases of industrial interest. In this paper, Aspergillus flavus lipase (AFL) was isolated from the traditional tannery of Fez city in Morocco; it kept its stability even in the presence of high concentrations of detergent from 0 to 10 mM sodium deoxycholate (NaDC). Bile salts showed no inhibitory effect on the lipolytic activity, whereas the calcium salts showed a stimulating action on the lipase activity. Unlike most of the lipases which were quickly denatured at the lipid/water interface, the accumulation of free fatty acids at the oil/water interface did not affect the activity of the enzyme which effectively hydrolyzed the emulsified olive oil even in the absence of bile salts. Furthermore, AFL was more active on long chain triacylglycerols than on short chain triacylglycerols. This study allowed us to prove that AFL had the interfacial activation phenomenon. A 3D structure model of AFL was built and we have concluded that the ratio hydrophobic surface/hydrophilic surface was 51% versus 50%; it could be responsible for a higher tolerance to the presence of free fatty acids at lipid/water interface.
Triacylglycerol acyl hydrolase (EC.3.1.1.3), lipases belong to the carboxylic ester hydrolases family. The physiological role of lipases is to hydrolyze triglycerides to diglycerides, monoglycerides, glycerol and fatty acids (Mats and Karl, 1994). These enzymes exist in all living organisms; they have the ability to achieve synthesis reactions such as esterification (reaction between an acid and an alcohol), the transesterification (ester andalcohol), interesterification (ester and ester), and in transfer reactions acetyl group of an ester. Lipolytic enzymes are perfectly soluble in water.
They are responsible for the hydrolysis of lipids which act on insoluble lipid substrates in water (Bénarouche et al., 2014). Lipids are formed by an aliphatic backbone, which are cyclic or polycyclic, that constitute the hydrophobic portion, which can be fixed polar groups and form the hydrophilic portion. Lipids include fats, oils, waxes and certain substances that are related (sterols, steroids, terpenes, etc).
The study of lipases has contributed to the development of the interfacial enzymology, which catalysis occurs in a heterogeneous medium at oil-water interface. The bio-chemical properties of these enzymes depend much on the quality of the interface and certain conventional parameters such as pH or ionic strength (Bénarouche et al., 2014). Lipases and esterases are important biocatalysts and particularly suitable for industrial applications, as they are very stable and active in organic solvents (Schmidt et al., 2004).
Chemicals
Tributyrin (99%; puriss) and benzamidine were from Fluka (Buchs, Switzerland); tripropionin (99%, GC) was from Jansen (Pantin, France); phosphatidylcholine, sodium deoxycholic acid (NaDC), Tween 20, yeast extract and ethylene diamine tetraacetic acid (EDTA) were from Sigma Chemical (St. Louis, USA); gum arabic was from Mayaud Baker LTD (Dagenham, United Kingdom); acrylamide and electrophoresis grade were from BDH (Poole, United Kingdom); PVDF membrane was purchased from Applied Biosystems (Roissy, France); casein peptone was from Merck (Darmstadt, Germany); and pH-stat was from Metrohm (Switzerland).
Lipase activity determination
The lipase activity was measured titrimetrically at pH 8.5 and 40°C with a pH-stat under standard conditions using TC4 (tributyrin) (0.25 ml) in 30 ml of 2.5 mM Tris–HCl pH 8.2, 2 mM CaCl2, 2 mM NaDC (sodium deoxycholate) or olive oil emulsion (10 ml in 20 ml of 2.5 mM Tris–HCl pH 8.2, 2 mM CaCl2, 4 mM NaDC); the olive oil emulsion was obtained by mixing (in a Waring blender) 10 ml of olive oil in 90 ml of 10% GA (gum Arabic) as substrate (Sarda and Desnuelle, 1958). Some lipase assays were performed in the presence of bile salts. Assays were carried out in 30 ml of 2.5 mM Tris–HCl buffer pH 7.0 containing 0.1 M NaCl. Standard conditions for measuring enzyme activity at increasing esters concentrations have been described previously (Stöcklein, 1993). When measuring AFL activity in the absence of CaCl2, we added EDTA to the lipolytic system. Lipolytic activity was expressed as units. One unit corres-ponds to 1 µmol of fatty acid released per minute.
pH-stat titrimetric assay
Titration method pH-stat of the liberated fatty acids from triacyl-glycerol continuously stirred, and coupled to an automatic burette, recorder and connected to a circulating water bath. This technique allows for automatical titration of the fatty acids released by addition of 0.1 N sodium hydroxide, maintaining the reaction medium at a constant pH during the hydrolysis of triacylglycerol emulsion by a lipase, and permits in varying the assay conditions, including sub-strates and bile salts. The pH chosen is generally appropriate in the optimum pH of the enzyme studied (Taylor, 1985). The enzyme activity is expressed in International Units (1U = 1μmole of fatty acid released / min).
Determination of protein concentration
Protein concentration was determined as described by Bradford (1976) using BSA (E1% 1cm = 6.7).
Procedure of AFL purification
1000 ml of culture medium, obtained after 72 h of cultivation, were centrifuged for 15 min at 8500 rpm and filtered to remove the cells. The supernatant containing extracellular lipase was used as the crude enzyme preparation.
Ammonium sulphate precipitation
The cell-free culture supernatant was precipitated using solid ammo-nium sulphate to 70% saturation. The pellet obtained after centri-fugation (30 min at 8500 rpm) was dissolved in 10 ml of buffer A (20 mM sodium acetate pH 5. 4, 20 mM NaCl, and 2 mM benzamidine) and within 5 min, insoluble material was removed by centrifugation at 13,000 rpm.
Heat treatment
The supernatant obtained (10 ml) was incubated for 30 min at 70°C and in 5 min; insoluble material was removed by centrifugation at 13,000 rpm. Filtration on Sephadex G-100: The enzyme solution (10 ml) was applied to a Sephadex G-100 column (3 ×100 cm) previously equilibrated in buffer A at a rate of 30 ml/h. The fractions containing the lipase activity (eluted at a void volume) were pooled.
Cation exchange chromatography
Active fractions eluted from Sephadex G-100 column were poured into a mono-S Sepharose cation exchanger equilibrated in buffer A. The column (2 ×30 cm) was rinsed with 400 ml of the same buffer. No lipase activity was detected in the washing flow. Adsorbed material was eluted with a linear NaCl gradient (300 ml of 20 to 500 mM in buffer A) at a rate of 45 ml/h. AFL activity was eluted between 170 and 220 mM NaCl. The fractions containing the lipase activity were pooled and concentrated.
Effect of free fatty acids
The lipase activity was measured according to various substrate (TC4, olive oil) in assigned concentrations ranging from 0-40 mM. The Michaelis-Menten (KMapp) and the maximum velocity (Vmax) for the reaction were calculated by Lineweaver-Burk (Taylor, 1985).
Effect of detergents
The lipase activity was measured using tributyrin and olive oil as a substrate in the presence of increasing concentrations of NaDC ranging from 0 to 10 mM, under optimum conditions of pH and temperature (Taylor, 1985).
Effect of calcium
The lipase activity was measured using tributyrin and olive oil as a substrate in the presence of increasing concentrations of calcium from 0 to 6 mM and under optimum conditions of pH and temperature. In the absence of calcium, the lipase activity is measured in the presence of 10 mM EDTA or EGTA (Taylor, 1985).
3D structure prediction
The AFL structure was modelled using the 3D coordinates of the closed form of the Aspergillus lipases (AL) (PDB code 3DXL_A). The method of minimization (Gromos96) (located in the server Swiss-PDB-Viewer) was used with the version of force field GROMOS43B1. This method enabled the evaluation of minimizing energy of the three dimensional structure and the distortion compensation. The geometry quality of the model was checked using PROCHECK program (Laskowski et al., 1993).
Production of lipase
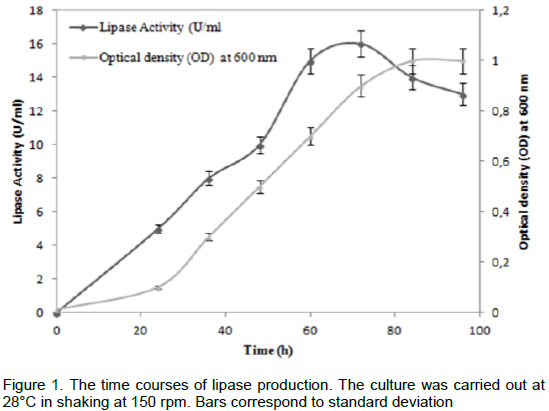
The maximal production of AF lipase with inoculum size of 3 × 107 cells/ml was 16 U/mL, which was obtained by incubating 1 ml of the enzyme with olive oil emulsion (10 ml in 20 ml of 2.5 mM Tris-HCl pH 8.2, 2 mM CaCl2, 4 mM NaDC) as substrate, The lipase activity was measured titrimetrically at pH 8.5 and 40°C with a pH-stat. The time course of lipase production was followed at 28°C with cell growth (Figure 1). The lipase activity was observed to start after incubation and reached the maximum (16 U/mL) at the end of the exponential phase corresponding to 72 h of cultivation.
Purification of AFL
The AFL was purified according to the procedure described. The protein elution profile which was obtained at the final step of the purification was shown in (Figure 2A).The purified lipase was homogeneous when tested by the Coomassie blue staining in SDS-PAGE (Figure 2). This figure shows that one band was revealed for AFL with a molecular mass of 55 kDa. The specific activity of the pure lipase reached 4300 U/mg using olive oil emulsified with gum Arabic as substrate at pH 8.5 and 40°C. Under the same conditions, a specific activity of 3400 U/mg was obtained when using TC4 as substrate.
Kinetic studies of AFL
Lipase hydrolysis emulsified triglycerides in the presence of bile salts. Some microbial lipases like Rhizopus oryzae lipase (Ben Salah et al., 1994) may lose its enzymatic activity, when TC4 or olive oil is used as substrate in the
absence of amphipathic reagent; the high energy existing at lipid/water interface is responsible for their irreversible denaturation. The enzyme denaturation cannot be reflected in the number of disulfide bridges, but behave very differently at interfaces. AFL was able to hydrolyze the TC4 or the olive oil emulsion alone (Figure 3). The kinetics of substrate hydrolysis remained linear for more than 20 min, accordingly; AFL probably presented a three-dimen-sional structure allowing it to hydrolyse its substrate efficiently and without any denaturation at high interfacial energy. Also, it tolerated the presence of long-chain free fatty acids, at the olive oil/water interface without any addition of amphipathic reagent (NaDC, Triton X-100); a difference between Aspergillus lipase and other microbial lipases which had a strong preference for short-chain substrates (Simons et al., 1996; Laachari et al., 2013; Sayari et al., 2001).
Effect of calcium on AFL activity
Metal cations, particularly Ca2+, plays important role in influencing the structure and function of lipases (Shangguan et al., 2011). The activity of lipases may depend on the presence of Ca2+ ions, like the staphylococcal lipases (Elkhattabi et al., 2003). The effect of various Ca2+ concentrations on the rate of hydrolysis of AFL was studied. A specific activity of 4300 U/mg was measured in the presence of 10 mM of chelator such as EDTA or EGTA, when using olive oil emulsion as substrate. In the absence of chelators, the specific activity of AFL reached 4900 U/mg at 2 mM CaCl2 (Figure 4). The enzymatic activity of AFL was stimulated by Ca2+. The lipases from P. Glumae and S. hyicus (Elkhattabi et al., 2003; Tiesinga et al., 2007) contained a Ca2+ binding site which was formed by two conserved aspartic acid residues near the active-site, dramatically enhanced the activities of these enzymes.
Effect of detergents
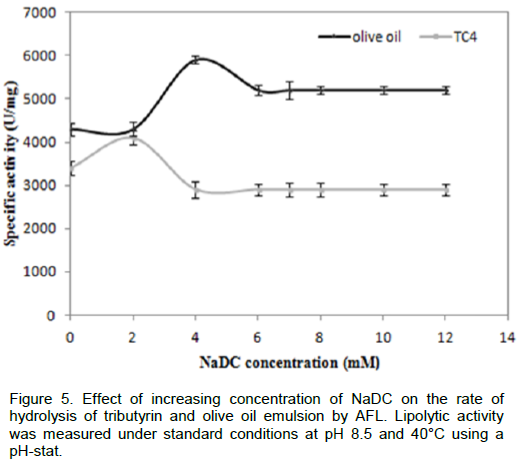
To verify whether AFL was capable to hydrolyze triglycerides in the presence of surfactants such as bile salts, we measured the rate of hydrolysis of TC4 and emulsified olive oil in the presence of various NaDC concentrations, from the results as shown in Figure 5, we notice that the NaDC has no inhibitory effect on activity of lipolytic enzymes even at a high concentration (10 mM). This result confirmed that unlike many lipases, such as Staphylococcus xylosus lipase, the maximal activity was reached in presence of 2 mM of NaDC using tributyrin as substrate. In presence of 4mM of NaDC, only 40% of residual activity was detected (Mosbah, et al., 2005). AFL was able to reach its substrate even in the presence of certain active agents surface, such as bile salts. Similar results were obtained with SSL (lipase from Staphylococcus simulans) (Sayari et al., 2001). Similarly, SSL was not inhibited by anionic detergents such as NaDC (Simons et al., 1997). Therefore, it can be inferred that probably AFL had a higher penetrating power than those of other microbial lipases that enabled it to hydrolyze the olive oil or TC4 in the presence of bile salts.
3D structure model of AFL
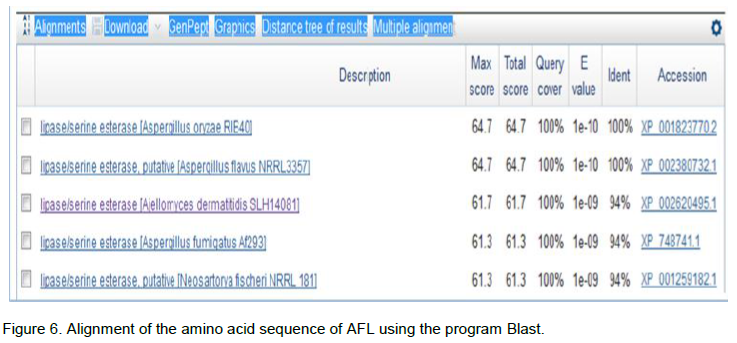
The research of homologous with the AFL sequence was made in the database using the BLASTp (Basic Local Alignment Search Tool protein) program by multiple alignment and the results are shown in Figure 6. Sequence analysis allowed us to reveal a 100% homology with A. flavus NRRL3357 (33.5 kDa), A. oryzae RIB40 and Ajellomyces dermatitidis SLH14081. In order to analyze sequences in databases NPSA (network protein sequence analysis) that allowed the comparison of protein sequences, a sequence alignment of the N-terminal portion of AFL was completed.
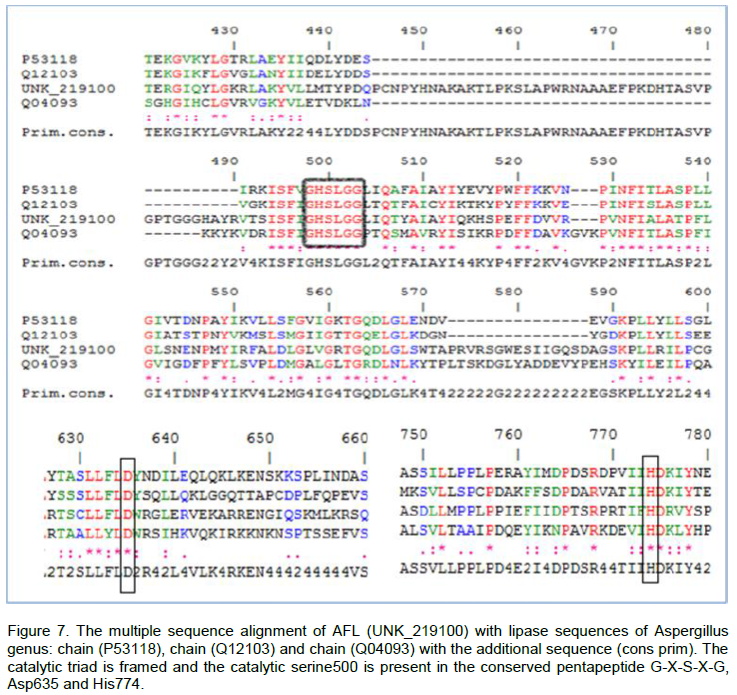
From Figure 7, we observe that the catalytic triad of AFL constituted Ser500, Asp635 and His774. The three amino acids were located on the C-terminal side of β core lamina. We also noticed that the Ser500 was a part of the pentapeptide Gly-X-Ser-X-Gly and located at the apex of the nucleophilic elbow between strand β5 and helix α5. The alignment of 3 sequences structures with the reference sequence allowed us to identify the location of the catalytic triad of AFL (Figure 7). Indeed, the catalytic triad (Ser-His-Asp) was a characteristic structure of a well known serine proteases (Brumlik and Buckley, 1996). It was also observed in the catalytic sites in several lipases with a carboxylate residue of aspartic acid or glutamic acid. These amino acids were generally determined by chemical modification studies (Ruiz et al., 2007) or by site directed mutagenesis (Hyun-Ju et al., 2000). The model of the Bacillus stearothermophilus lipase P1 constructed by the use of the basis of secondary structure predictions, showed an organization in fold and this enzyme α/β-hydrolase was identified by the catalytic triad of Ser-113, Asp-317 and His-358, in close proximity to each other which played a key role in the catalytic mechanism (Sinchaikul et al., 2001).
In order to create a sample of the closed form of the A. flavus lipase, we used the automatic modeling by the Swiss-Model server (http://www.expasy.org/spdvb). The model of AFL was a subject of several cycles of energy minimization using Gromos96 installed on the server Swiss PDB. The superposition of the two closed structures gave an average standard deviation (rmsd) equal to 1.46 Å. The stereochemical quality of the closed model of AFL and statistical analysis of the distribution of amino acids in the Ramachandran plot were also tested by the program PROCHECK (Laskowski et al., 1993). According to the Ramachandran plot (Figure 8), we found that 99 and 97% of non-polar and polar amino acids respectively, were located in suitable areas (Figure 8).
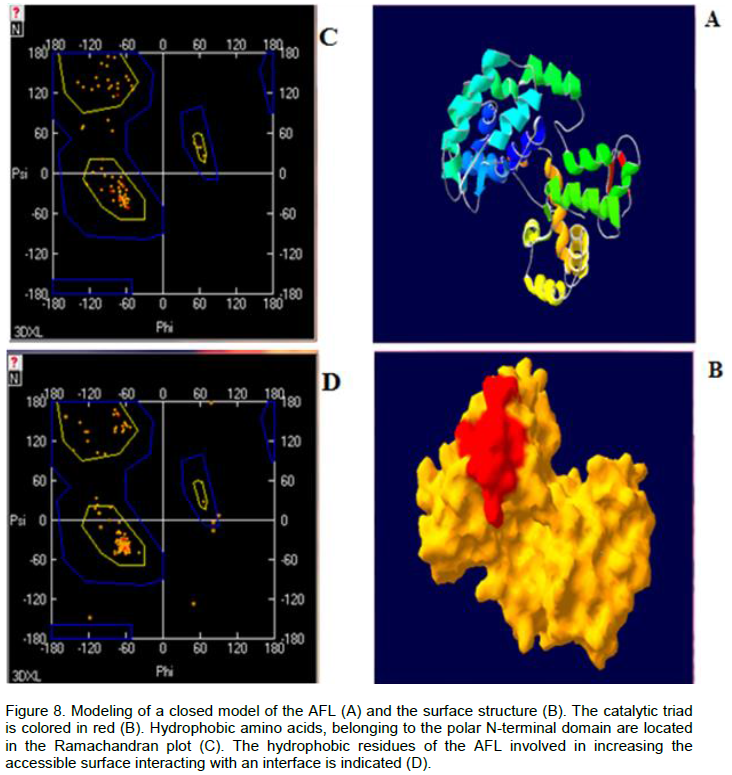
The crystal structures of Bacillus sp. H257 lipase in its free form at 1.2 was found complexed with phenylmethyl-sulfonyl fluoride at 1.8 Å. The catalytic residues were buried at the bottom of a long chain ~ 22 Å, from the surface to the active site, the bottom of the binding pocket of the substrate had more polar character with contributions from catalytic residue Ser97, His226, Met98 and Phe29, built the oxyanion hole, which stabilized the tetrahedral intermediate formed during the hydrolysis reaction (Rengachari et al., 2012; Laachari et al., 2014). Furthermore, the study of the total surface accessible (N-terminal domains) of the closed form of AFL, showed that the ratio hydrophobic surface/hydrophilic surface was 51% versus 50%. This could explain the fact that AFL tolerates accumulation of long chain fatty acids in the lipid/water interface and had a linear kinetics over 15 min, during the hydrolysis of an emulsion of oil olive.
The structure of the lipase from A. niger was composed of a core domain (residues 1-82 and 97-269), showing the typical characteristics of the α/β hydrolase and a lid domain (residues 83-96), with a simple pattern of loop-helix-loop. The orientations and the positions of residues of the catalytic triad (Ser145-Asp198-His260) followed the movement of the lid (Liu et al., 2013). These observations allowed us to suggest that the structure of the lipase from A. flavus conformed to all the characteristics of the lipases in the open conformation.
The interest of microbial lipases in biotechnological applications has taken a meteoric rise in recent years. Therefore, the industry requires new enzymes, which meet the criteria of use, particularly in terms of thermostability. The A. flavus lipase kept their stability even in the presence of high concentrations of detergent (NaDC). Calcium salts showed a stimulating action of the lipase activity. Molecular modeling of the 3D structure was also carried out and the results have allowed us to demonstrate that AFL tolerates the accumulation of long chain fatty acids in the lipid / water interface. These results show that AFL has biochemical properties attractive for various industrial applications.
The authors did not declare any conflict of interest.