ABSTRACT
Bacterial communities are actively involved in composting process but the environment within the compost influences their diversity, abundance and succession. In this study, the authors investigated the bacterial communities in tropical crop waste compost using pyrosequencing. Municipal crop wastes from the tropics (Uganda) were composted under four different low-technology methods. Samples were collected from the early thermophilic, late thermophilic, and mesophilic phases, and from mature compost. Pyrosequencing of the amplified variable V4 region of the 16s rDNA generated over 110 000 sequences. Chao1 and cluster analysis at 3% dissimilarity showed that bacterial community richness declined during the composting process. The community was dominated by a few bacterial taxa during the thermophilic phases. Species evenness increased as compost progressed to maturity despite a decline in the number of taxa over the successional progression. Bacterial community diversity, abundance and succession changed with the composting method. This pattern of diversity may be attributed to competition and selection during the microbial succession. A total of 22 phyla and 513 genera were identified from all the methods in the entire composting process. The most abundant phyla were Proteobacteria, Firmicutes, Bacteriodes and Actinobacteria. Pyrosequencing provided more information on compost bacterial community diversity and abundance than previously used molecular methods. Several novel bacteria existing in tropical crop waste compost remained unclassified.
Key words: 16S rRNA, succession, compost, bacterial community, pyrosequencing.
The composting process is primarily driven by micro-organisms and understanding their succession, ecology, characteristics, diversity and abundance is important for its optimization as well as the understanding of nutrient cycling. Microorganisms involved in composting are primarily bacteria and fungi (Riddech et al., 2002), which have variable roles, functions and ability. Therefore, revealing the identity, nature and functions of the organisms is a prerequisite for determining pathways of the composting process and the quality of mature compost. Composting has been widely used in management of organic wastes. In Uganda, about 5000 metric tons of municipal solid waste (MSW) was generated in Kampala city alone per month, which can be composted. MSW compost has a long residual effect in soil than only animal waste compost (Zing et al., 2017). The quantity of waste generated in the urban is increasing with population. Han et al. (2018) reported a positive linear relationship between waste generation amount and population size (R2 = 0.9405). Most of the MSW composting studies in the tropics have concentrated on physicochemical properties of the process and product but there is relatively little knowledge available on microbial characteristics. A high throughput method like pyrosequencing would be more suitable to elucidate the microbial community during MSW composting. Pyrosequencing provides massive parallel sequence based taxon identification compared with previously used molecular techniques (Shendure and Ji, 2009; MacLean et al., 2009; Fullwood et al., 2009). It has revealed more bacterial diversity in soil (Roesch et al., 2007; Acosta-Martínez et al., 2008; Jones et al., 2009) and during composting (de Gannets et al., 2013). Therefore, it is plausible that higher bacterial richness in compost than currently known will be revealed through pyrosequencing. The diversity, abundance and succession of bacteria community in the tropical mixed wastes such as municipal crop wastes composting ecology is currently to a large extent unexplored. The composting process can be divided into four phases: initial mesophilic phase (<40°C), followed by a thermophilic phase (>40°C) before dropping to a second mesophilic phase and finally the compost maturing phase. Bacterial and fungal communities were noted to dynamically relate to the functional composting progress phases (Xi et al., 2016; Zhao et al., 2016). Each of these phases is characterized by different compost ecological conditions such as temperature, pH and nutrient concentrations, which may affect microbial community structure. Bacteria use a broad range of enzymes to degrade available organic substrates such as soluble carbohydrates, proteins and complex lignin, chitin, polyphenols and cellulose.
Although, there have been studies on bacteria community in composting elsewhere, few of these studies have traced and described the microbial community during an entire composting process. For example, Takaku et al. (2006) used DGGE and PCR amplified 16S rDNA to study microbial community during composting of food garbage and rice hulls in Japan. Results showed change in microbial community between composting stages. A total of 33 DGGE bands which were identified belong to phyla Proteobacteria, Firmicutes, Bacteroidetes and Actinomycetes. In a related study, high diversity of Formicates and Bacteroidetes was observed during thermophilic phased during composting of solid agricultural wastes (Song et al., 2014). Culture-method was noted to underestimate the diversity of microorganisms during the composting cycles as compared to sequencing of 16S rRNA (Chandna et al., 2013). Earlier, Horisawa et al. (2008) determined bacteria succession during decomposition of garbage but in a batch-fed garbage decomposer and showed that microbial community in compost were similar at each stage of composting irrespective of the environmental conditions. Taiwo and Oso (2004) and Rebollido et al. (2008) traced microbial succession during composting of organic municipal solid wastes in Nigeria and Cuba, respectively but used the culture methods. There was a higher community of bacteria followed by Actinobacteria and fungi during composting dominated by Bacillus, Streptomyces, Actinomyces, Pseudomonasi and Azospirillum isolates (Rebollido et al., 2008). Bacillus, Streptomycetes and Pseudomonas in addition to faecal coliforms, Seratia and Proteus were the dominant microbial community isolates from pit method composted urban organic wastes (Taiwo and Oso, 2004). Other studies to establish bacteria community during composting process used kitchen waste, sewage sludge and source-separated municipal solid or household wastes mainly in temperate climate (Kowalchuk et al., 1999; Peters et al., 2000; Ryckeboer et al., 2003; Connon et al., 2005; Neher et al., 2013; Song et al., 2014). Some studies have concentrated on microbial succession within one phase of the compost process. For example, Danon et al. (2008) studies bacterial community succession in curing compost of sewage sludge and yard wastes from a commercial composting facility in Israel and reported microbial community changes but the dominant phyla were Proteobacteria, Bacteroidetes and Actinobacteria. The objectives of this study were to explore the bacterial community diversity, abundance and succession during composting of tropical MSW in four low-technology composting methods using pyrosequencing of the 16S rDNA V4 region.
MSW were obtained from a fresh food market in Kampala city, Uganda and composted at Makerere University Agricultural Research Institute, Kabanyolo (MUARIK). MUARIK is about 17 km north of Kampala City. The final composition of the MSW after removal of non-biodegradable materials and non-plant materials, included banana residues, fruits (e.g. oranges, mangoes, avocadoes, papaws, water melon and jackfruits). Other constituents were vegetables (cabbages, tomatoes and egg plant), sweet potato vines, maize residues, beans and peas haulms. The crop wastes were manually homogenized before composting.
Study design
Composting was carried out in open field experiments where four low technology methods (treatments) were arranged in a randomised complete block design, each with three replicates. The methods were: (i) pit-open (PO); (ii) pit-covered (PC); (iii) above ground-open (AO); (iv) above ground-covered (AC). Each of the composting structures had dimensions of 2 x 1.5 x 1.6 m (length x width x depth/height). The composting materials were turned manually with hands and forked hoes twice each week for the first three weeks, then once a week for another two weeks and once a fortnight for another four weeks in order to increase aeration and subject all the material to high temperatures, as well as mix the materials during the composting process. Moisture was monitored gravimetrically each week from a composite sample of compost and moisture content was maintained between 40 and 60% for the first 35 days.
Laboratory analysis
Compost samples were collected on each day of turning. A compost sample for each replicate comprised of samples picked randomly from ten spots during turning. The samples were packed in 200-ml plastic vials, transported under frozen condition and stored at -80°C until used for analysis. Microbial molecular studies were done at the Swedish University of Agricultural Science (SLU) in Sweden. Genomic DNA from the frozen compost was extracted following procedures using 3% hexadecyltrimethylammonium bromide (CTAB) buffer incubated at 65°C. Thereafter, centrifuge was done at 10000 round per minutes (rpm) for 5 min and the top 200 μl was extracted and mixed with chloroform and centrifuge was done again at 10000 rpm. This was followed by extraction of the top 150 μl and mixed with cold isopropanol and kept on ice for 30 min and later centrifuge at 13000 rpm for 13 min. Taking care not to dislodge the DNA pellets, the supernatant was discarded. The DNA pellets were washed with 200 μl of cold ethanol (70%) and centrifuge at 6500 rpm for 5 min. The supernatant was again discarded and pellets dried on the bench and eluted with 200 μl milliQ water and stored at -26°C until used. The quality of the DNA was very low and could not be used for PCR steps. Therefore, its quality was further enhanced by purification using a Jet Quick kit (Genomed GmbH, Löhne, Germany) following the manufacturer’s instructions and eluted in 50 μl of preheated TE buffer. The concentration and quality of DNA was measured on the NanoDrop Spectrophotometer (ND1000). Purified DNA was kept at -26°C until used. Nested PCR was used for pyrosequencing of V4 16S rDNA. Universal bacteria primers 27F (5' AGAGTTTGATCMTGGCTCAG 3') and 907R (5-CCGTCAATTCMTTTRAGTTT-3) were used for the first PCR step. A PCR reaction volume of 20 μl contained: 1x buffer, 0.2 μM dNTPs, 0.2 μM 27f and 907R each, 0.75 mM of MgCl2, 0.02 U μl-1 red Taq polymerase and 100 ngμl-1 DNA was used. Amplification was performed on Applied Biosystems 2720 Cycler with initial denaturing at 94°C for 3 min, followed by 25 cycles of denaturation (92°C for 45 s), annealing (50°C for 30 s) and extension (72°C for 30 s). There was a final extension for 7 min at 72°C and then maintained at 4°C. The PCR product integrity was checked on 1% gel. In the second PCR, primers V4R1-4 with pyrosequencing adaptor-A added in the 5' end and V4F with tag and pyrosequencing adaptor B added in the 5' end were used (Cole et al., 2009). For the second PCR, 1 µl of the first PCR products were used as templates with the following: 1x buffer, 0.08 μM dNTPs, 0.38 mM MgCl2, 0.15 μM AV4R, 0.08 U/μl red taq polymerase and 0.15 μM V4F primer, different tag for each sample in a total PCR reaction volume of 50 μl. The samples were gel purified using QIA Quick gel extraction kit, was concentration measured with a spectrophotometer before pyrosequencing on a 454 GS FLX instrument at a concentration of 10 ng/µl. The sequences have been deposited in the Short Reads Archives (SRA) at NCBI (under accession number SRA 009487.3).
Alignment and clustering of 16S rDNA fragment
The sequences were processed using Ribosomal database project (RDP), release 10 pyrosequencing pipeline (Cole et al., 2008). Using the initial pipeline process, raw reads were sorted for each sample using the used tagged. This was followed by trimming off tags and eliminating poor and shorter sequences less than 150 bp. The same procedures were followed to eliminate sequences shorter than 200 bp. The libraries with sequences > 200 bp were used for classification studies and diversity studies. RDP using the naive Bayesian classifier classified more accurately sequences ≥ 200 bp (Wang et al., 2007). The tag-trimmed sequences were aligned by (i) methods of composting and (ii) composting phases and then clustered by complete linkage at 20 with a 5% increment and 3%with 1% increment. Clusters at 3% dissimilarity were used to estimate the number of OTUs in previous studies with pyrosequencing (Roesch et al., 2007; Acosta-Martínez et al., 2008). The clusters were therefore used at 3% dissimilarity to estimate bacterial community richness and diversity with the parametric indicator rarefaction and non-parametric indices (Shannon and Chao1) using the RDP pipeline. The rarefaction curves were drawn in MS excel software and curve fitting was performed with GraphPad Prism 5.0 (GraphPad Software) at 95% confidence interval using two phase association model. From previous studies, Roesch et al. (2007) observed that it is necessary to model and extrapolate rarefactions curves or use non-parametric methods to estimate OTU richness taking into account the community structure. The RDP classifier tool was used to assign the sequences to phylotypes based on Naive Bayesian classifier of 16S rRNA. The RDP library was used to compare library sequences between methods of composting. Non-metric multi-dimensional scaling (NMDS) was used for ordination of clusters at the 3% level as cluster data are categorical and not normally distributed. Data were square-root transformed and the Bray-Curtis dissimilarity index was used as input as implemented in Vegan. Environmental variables (pH, temperature, total nitrogen, total carbon, NO3-, NH4+) were overlaid to the ordination by means of vector-fitting. Significance was achieved by means of permuting data 999 times.
Bacteria community succession between phases of composting
A total of 110051 sequences were obtained and when filtered for sequences > 200 bp, the number reduced to 109728 of which 38243, 37372, 25641 and 8474 were from early thermophilic, late thermophilic, second mesophilic phases and mature compost respectively. RDP classified 99.7% of the sequences as bacteria at 80% bootstrap. Reducing the confidence level of 70%, RDP classified 99.9% of the sequences as bacteria leaving 40 sequences belonging to Archaea. Thirty five of the Archaea sequences were from thermophilic phase while four sequences and one sequence were from second mesophilic phase and mature compost respectively. Rarefaction curves showed variations in the number of OTUs at different phases of composting (Figure 1). From curve-fitting of rarefaction curves, 6919 OTUs (6884 and 6948 lower and upper 95% confidence interval respectively) were estimated during early thermophilic phase. The late thermophilic had 5141 OTUs (5118 to 5164 lower and upper 95%), while the mesophilic phase had 5118 OTUs (5093 to 5142 lower and upper 95% confidence interval) and further declined in mature compost to 2895 OTUs (2881 to 2909 lower and upper 95% confidence interval). This trend was similar to the cluster and observed OTUs and Chao 1 estimations at 3% dissimilarity. Shannon index showed a decrease in species richness between early thermophilic phase and mature compost phases (Table 1).
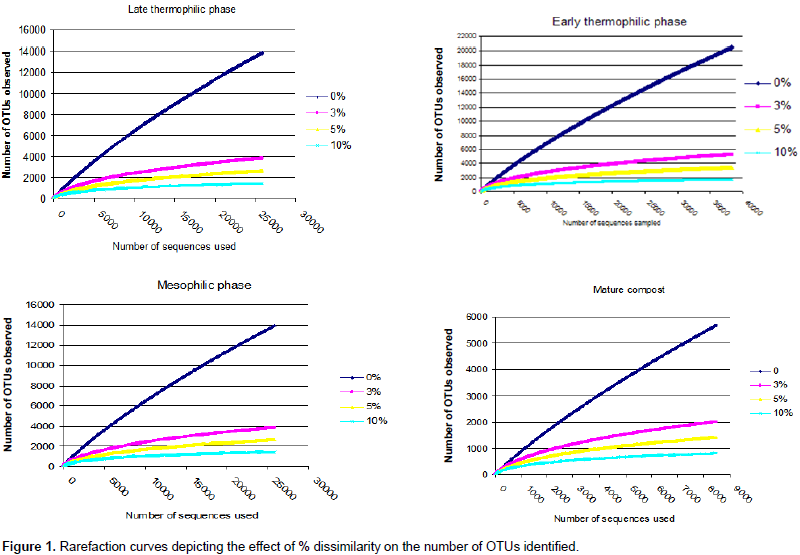
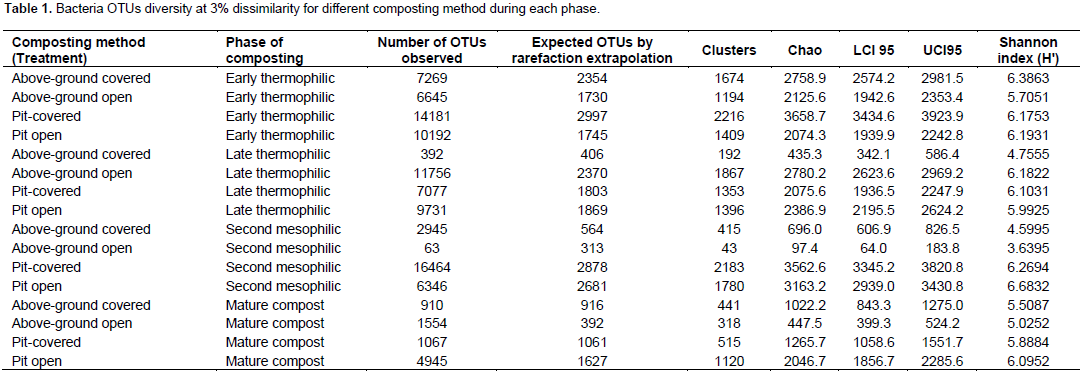
The pit methods generally had the highest richness. The PC method had the highest bacteria diversity as shown by OTUs estimated by rarefaction extrapolation in the early and mesophilic phases. It was followed by the PO method, which had the highest rare faction estimated OTUs in mature compost. On the other hand, AO method had the least bacteria community diversity in the entire composting process except in the late thermophilic phase (Table 1). The clusters and Chao 1 values followed similar pattern except in the early thermophilic phase. Species richness decreased as the evenness increased during composting in all methods as shown by the H' index (Table 1). The AO method had successively increased bacterial diversity as compared to the other methods as compost progressed through maturity.
Proteobacteria, Firmicutes, Bacteriodetes, Actinobacteria and Planctomycetes were the most abundant bacterial phyla throughout the composting process (Table 2). Proteobacteria community abundance was highest during early thermophilic phase, decreased in late thermophilic and mesophilic phases but later increased in mature compost. A similar pattern was observed for Cyanobacteria, Nitrospira, Spirochaetes and Acidobacteria. The abundance of Bacteriodetes increased from the early thermophilic through the mesophilic phases but decreased in mature compost. There wasa similar pattern for TM7, Gemmitimonadates, Deinococcus-Thermus, Chlamydiae and Chloroflexi. The Actinobacteria community increased abundance from 5.1% during thermophilic phase to 8.0% in late thermophilic phase but thereafter decreased successively. Planctomycetes community abundance was nearly consistent during early and late thermophilic phases but later successively increased through compost maturity. The minor phyla (Chlamydiae, Deferribacteres, Chloroflexi, Acidobacteria, Lentisphaerae, Tenericutes, TM7, BRC1, OP1, OD1, Verrucomicrobia, Gemmatimonadates, Deinococcus-Thermus and Spirochaetes) contributed less than 10% to the phyla abundance during all phases of composting. Several sequences remained unclassified with the highest proportions at 38.5 and 26.8% of the sequences from second mesophilic and late thermophilic phases, respectively (Table 2).
At class level, α-proteobacteria and γ-proteobacteria had the highest abundance during the composting process followed by Bacteroidetes except in mature compost (Table 3). Planctomycetacia successively increased in abundance and dominance during composting until mature compost. Chloroflexi, Lentisphaerae and Deferribacteres were eliminated before compost matured (Table 3). At genus level, Aquicella, Petrimonas, Alkaliflexus, Brevibacterium, Phenylobacterium were the most abundant genera from early thermophilic to mesophilic phases but were succeeded by Roseomonas, Streptococcus and Brevundimonas in mature compost (Table 4). Genera diversity successively decreased during composting. There were 246 genera during early thermophilic, which decreased to 201 in late thermophilic phase, further decreased to 197 and 150 for second mesophilic and mature compost respectively from all composting methods combined. There were some genera which appeared only in one composting phase. There were 44, 23, 21 and 22 unique genera that appeared only in the early thermophilic, late thermophilic, second mesophilic phases and mature compost, respectively. However, from the 20 most abundant genera (Table 4), Crypanaerobacter was detected only in early thermophilic phase, Pirellula was eliminated after the second mesophilic phases, while Plastopirellula and Veilonella were present only in mature compost. There was a decreasing successional trend in the number classes and genera for all the composting methods. More bacterial classes and genera were observed during early thermophilic and richness decreased in mature compost. In the early thermophilic phase, the AC method was significantly richer in the number of classes than all the other methods.
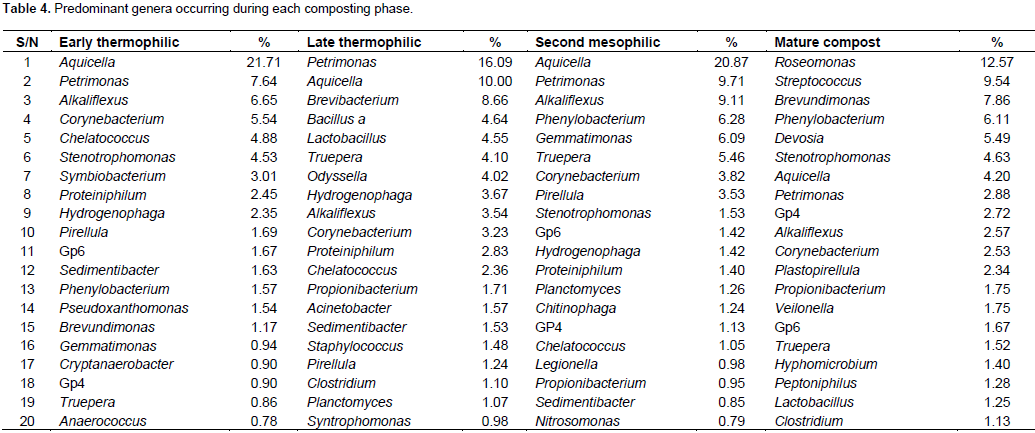
Progressing to late thermophilic phase, the AO method had the highest number of classes while the AC method declined considerably. In the second mesophilic phase, the PC method had the highest class richness while the AC method had the lowest number of classes observed in the entire composting process. In mature compost, the most and least number of classed were from PO and PC methods, respectively. A similar pattern of succession was observed even at genera level (Figure 2).There were differences in class or genera diversity and abundance among different composting methods even within the same composting phase. For example, during early thermophilic phase, the genus Aquicella was the most dominant in open systems (AO and PO methods) while Alkaliflexus and Corynebacterium were the most abundant in AC and PC methods, respectively. In the late thermophilic phase, the most abundant genera were Brevibacterium and Bacillus for the AC method, Petrimonas and Hydrogenophaga in the AO method, Petrimonas and Pirellula in the PC method, Aquicella and Petrimonas in the PO method. For the second mesophilic phase, the most abundant genera under different composting methods were Petrimonas and Corynebacterium in the AC method, Petrimonas and Alkaliflexus for AO method, Aquicella for the PC method, and Phenylobacterium and Gemmatimonas in the PO method. In the mature compost, the genera Terrimonas and Phenylobacterium were the most abundant in the AC method, Streptococcus, Stenotrophomonas and Corynbacterium in the AO method, Roseomonas, Phenylobacterium and Pirellula in the PC method and Roseomonas, Brevundimonas and Devosia in the PO method.
Bacteria succession in different composting methods
Bacterial succession patterns differed significantly depending on the composting methods and phases(p<0.01). For example, in the AC method (Table 2), there was a decrease in abundance of Firmicutes, Proteobacteria, Bacteroidetes and Planctomycetes between early thermophilic phase and mature compost.
However, Proteobacteria dominated the early thermophilic phase and mature compost while Firmicutes and Bacteroidetes were most abundant during late thermophilic and second mesophilic phases, respectively. The minor phyla abundance also successively increased during composting. The minor phyla included Cyanobacteria, Nitrospira, Deferribacteres, Chloroflexi, Chlamydiae, Acinobacteria, Lentisphaerae, Tenericutes, TM7, SR1, BRC1, Verrucomicrobia, Gemmatimonadates, Spirochaetes and Deinococcus-Thermus. For the AO method (Table 2), the abundance increased for Firmicutes and Actinobacteria while it decreased for Proteobacteria, Bacteroidetes and Planctomycetes. For the PC method (Table 2), Proteobacteria Bacteroidetes and Actinobacteria had a successive decline in abundance but it increased for Firmicutes and Planctomyces. There were notable changes in the minor phyla; Acidobacteria and Verrucomicrobia successively increased from 0.1 and 0.4 to 4.2 and 3.4% between early thermophilic phase and mature compost, respectively. For the bacterial succession under the PO method; there was a significant increase in abundance of Proteobacteria, Firmicutes and Planctomycetes and a decrease for Actinonacteria. The minor phyla Verrucomicrobia, TM7 and Acidobacteria were more abundant than Cyanobacteria, Chlamydiae, Chloroflexi, Lentisphaerae, Tenericutes and Gemmatimonadetes. NMDS analysis gave two convergent solutions after 13 trials, stress value 20.2 and non-linear fit R2 was 0.94 (Figure 3). Temperature, total organic carbon, NH4+ and NO3- explained most of the community variation but pH and total nitrogen were significant factors as well when vector-fitting was permuted 999 times (Table 5).
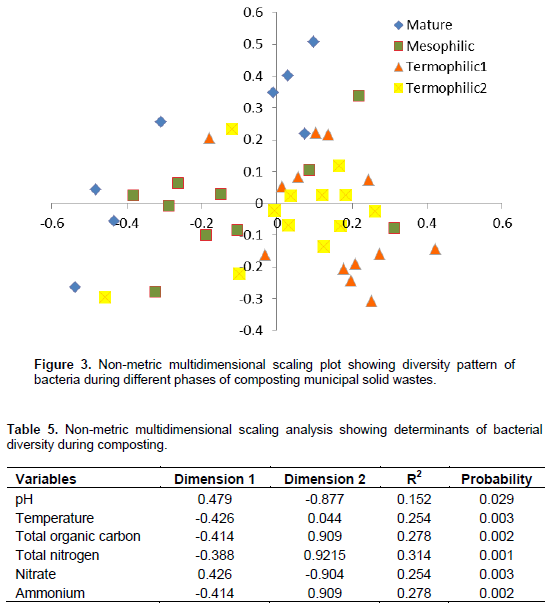
Pyrosequencing of 16S rDNA revealed successional patterns, decreased diversity of bacteria community and increased abundance of some taxa during composting than has been reported by any previously used molecular methods. Earlier, de Gannets et al. (2013) used 454 pyrosequencing and noted that diversity increased as composting progressed and matured compost was dominated by Archaea, Planctomycetes, Chloroflexi, α- and É£-protebacteria. A similar pattern was reported when source separated wastes were subjected to static aerobic composting where abundance of É£-protebacteria, actinobacteria and Bacillus was observed durng the composting cycle (Sundberg et al., 2011). The main microbial succession selection factors during composting include temperature, aeration, pH, moisture and nutrients. In this study, temperature, total organic carbon, NH4+ and NO3- explained most of the community variation (Table 5). The NMDS analysis separated samples after successional stages rather than according to methods (Figure 3). This is mainly an effect of temperature. Carbon was significantly lost during early stages of composting, here, it is shown that the loss of carbon is as well a significant driver of community shifts during the succession. The significant effect of NH4+ and NO3- on the community composition may show that nitrification is an important energy resource when the easily degraded carbon is depleted. The general trend of a decreasing number of taxa with an increasing evenness may be explained by shifts in microbial ecological strategies during the compost succession. In plant ecology, patterns of community distribution have been explained by competition, dispersal and resource utilization. Dispersal is important in the early succession after a disturbance whereas competitive species dominate in rich environments and stress tolerant species inhabit the environment when resources are depleted. At the initial compost phases, the substrates are rich in nutrients and are inhabited by a large number of microbial taxa.
As succession proceeds, the taxa show a more even distribution which is consistent with a shift to a community with many stress tolerant, less competitive species occurring in the late stages. Dispersal may be ofminor importance in the compost environment since the compost material already is colonized with a large number of micro-organisms. The observed dominance of a few bacterial taxa (Table 2) during the thermophilic phases may be the result of strong selective factors in combination with strong competitive ability (Song et al., 2014). In the early stage of composting, temperatures are high, thereby eliminating non-thermophilic micro-organisms. Nutrient availability or pH are unlikely to have restricted the number of taxa in the composts since a broad range of nutrients at high concentration were present in the initial compost and initial pH of the compost substrate was around neutral. It is therefore tempting to suggest that high temperature could be the main selection pressure for the dominance of the three genera: Aquicella, Petrimonas and Alkaliflexus in the early, late and early mesophilic phases. In an earlier study, Rebollido et al. (2008) observed that temperature was the most environmental factor which influenced microbial succession during composting process. Additionally, it is noted in the authors’ previous study (Tumuhairwe et al., 2009), that there were no significant differences in pH and nutrients among the composting methods except differences between composting phases. Those three genera are not well represented in GenBank with less than 50 sequences archived in the database. Furthermore, none of these genera has been reported to dominate compost communities before or specifically as thermophilic genera. In the future, it would be interesting to isolate representatives of these three genera to test their competitive ability against other isolates and their heat sensitivity in order to better understand why they dominate the initial phases of tropical municipal waste compost communities. In this context, it is interesting to add that most Archaea which include many well-known thermophiles (Lebedinsky et al., 2007; de Gannets et al., 2013) were found during the early thermophilic phase.
The lowest observed number of classes and genera from the AO method during second mesophilic phase (Figure 2) could be attributed to fewer numbers of good sequences obtained but would probably not have changed the observed decreasing community richness as shown by the rarefaction estimates. The successive increase in diversity of OTUs in the PO method as compared to the other methods may be because PO method provides most favorable condition for the bacteria community in contrast to the AO method. The most plausible cause of decreased bacteria diversity under the AO method could be moisture constraint. Temperatures are usually high in the tropics, which increases evaporation of moisture from compost when it is not covered. Proteobacteria, Bacteroidetes, Firmicutes, Actinobacteria and Planctomycetes which were the dominant phyla have been reported in other composting studies (Takaku et al., 2006; Danon et al., 2008; Székely et al., 2008; Vivas et al., 2009; Neher et al. 2013; Karadag et al., 2013; Song et al., 2014; Manyl-loh et al.. 2016). The high abundance and diversity of Proteobacteria was expected because this phylotype contains mainly chemo-organotrophs, which derive their energy from decomposition and oxidation of organic matter. The observation that Proteobacteria were most dominant during composting is consistent with composting studies elsewhere (Ntougias et al., 2006; Takaku et al., 2006; Danon et al., 2008; Vivas et al., 2009). However, the current results differ from the some previous studies by having α-proteobacteria as the most abundant class. Takaku et al. (2006) and Vivas et al. (2009) reported β-proteobacteria while Ntougias et al. (2006) observed γ-proteobacteria as the dominating classes in the composting process. The reason for this disparity is not clear but it may be attributed to differences in locality, composting substrate and methods used.
The high abundance of Bacteroidetes is not surprising as they are known to degrade macromolecules (Michel et al., 2002) which are prevalent in municipal waste compost. The decline in abundance of Bacteroidetes could be associated with the compost progressing to maturity when the macromolecules have been degraded. The genus, Stenotrophomonas which was ubiquitous in all composting phase together as well as genera Pseudoxanthomonas and Nitrospira contain denitrifying bacteria (Lipski and Altendorf, 1997; Chen et al., 2002) and could also have contributed to the significant loss of N during composting of municipal crop wastes. Planctomycetes have been reported in diverse ecosystems including compost (Buckley et al., 2006; Takaku et al., 2006; de Gannets et al. 2013). In compost, Planctomycetes have currently been detected during the early composting stages (Takaku et al., 2006). However, in this study, Planctomycetacia steadily increased from the seventh to third most abundant community between early thermophilic phase and mature compost (Table 2) as also reported by de Gannets et al. (2013). This contrasting observation could be due to the paucity of information on functional activity of Planctomycetes during composting. However, Planctomycetes have been reported to show chemo-litho-autotrophic growth through oxidation of ammonium (Strous et al., 1999) and could also contribute to nitrification. For the phylum Firmicutes, Clostridia dominated from early thermophilic through second mesophilic phases while Bacilli was more dominant in mature compost. Firmicutes have been reported during different stages of composting (Ntougias et al., 2006; Takaku et al., 2006; Karadag et al., 2013) and were favored by alkaline medium. Therefore, increase in abundance of Bacillus and Clostridia could be attributed to favorable alkaline pH during the composting. Characteristically, Bacilli and Clostridia are facultative and strict anaerobes, respectively.
Therefore, their presence suggests incomplete aeration during composting. Complete aeration may be difficult to achieve under the low-technology composting methods used in this study. However, there are reports of Bacillus and Clostridia from other composting studies (Ntougias et al., 2006; Takaku et al., 2006; Sundberg et al., 2011; Karadag et al., 2013; Fui et al., 2017 ) indicating that it is difficult to achieve complete aeration during composting. The most abundant genus within Firmicutes was Symbiobacterium which was detected among the predominant 20 genera during early thermophilic phase (Tables 3 and 4). The successional pattern of Symbiobacterium could be explained by high temperature during early thermophilic phase since it has previously been isolated from thermophilic compost (Ueda et al., 2001). Another abundant bacterial group was Actinobacteria (Table 3) predominated by Corynebacterium at the genus level (Table 4). Actinobacteria effectively degrade organic matter including complex substances such as cellulose and chitin and are favored by temperature between 45 and 55°C. Therefore, the successional pattern of Actinobacteria community during composting could be explained by the temperature gradient. The highest Actinobacteria community was observed during the late thermophilic phase when the temperatures were most favorable. The dominance of the genera Streptococcus, Plastopirellula and Veilonella communities in mature compost (Table 4) could be used as an indicator for compost maturity. Streptococcus had earlier been isolated from maturing compost from a mixed municipal solid waste and poultry manure in Nigeria (Taiwo and Oso, 2004). This could be the first study to identify the predominance of genus Devosia in mature compost, previously isolated from aquatic and soil environments (Nakagawa et al., 1996; Rivas et al., 2003). The presence of phyto-beneficial bacteria in mature compost improves its value for soil fertility management and such examples include Rhizobia and Bradyrhizobia for symbiotic nitrogen fixation and Roseomonas, a genus that includes plant-growth promoting bacteria. The current high number of taxa identified from compost than previous studies (de Gannets et al., 2013) further demonstrates the high throughput capacity of pyrosequencing in molecular microbial ecology studies even when several sequences remained unclassified at phylum and class levels. This study has also demonstrated the ability of pyrosequencing to detect both the dominant and minor bacterial population occurring during the composting process unlike PCR-DGGE or TRFLP which only detect the dominant bacteria taxa (Takaku et al., 2006).
The high number of unclassified bacteria sequences at each stage of composting, most of which belonged to either unknown or uncultured bacteria suggests that pyrosequencing is a powerful technique that provides details on microbial diversity within compost. It also highlights the presence of novel groups of unknown bacteria that may exists when composting MSW in the sub-Saharan tropical environment. This study revealed the presence of bacterial taxa belonging to OD1, OP10, BRC1, SR1, Deferribacteres, Lentisphaerae, Spirochaetes and TM7 phyla within a compost environment. To the authors’ knowledge, this is the first study to identify such high diversity and abundance of phyla, classes and genera existing in compost ecology in a single study. The significant variation in diversity and abundance of these minority phylotypes at different composting stages could provide insight into unknown functional groups that exist during the composting process. Although, all these genera were identified, their origin and functions during composting remain unknown. It could be that they were originally part of the municipal waste compost, introduced in water used for maintaining the moisture content during the composting process or drifted from soil and surrounding to composting material. Furthermore, it is also unknown whether they were inactive or in dormant state and this is one of the setbacks of DNA-based molecular techniques. However, the change in genotype abundance and diversity, and OTUs observed at different phases during composting period suggests an active community contributing to successional changes. In the future, it would be interesting to study functional aspects of microbial communities. This may be pursued via the study of functional genes that are involved in ammonium oxidation and the cultivation of representative taxa to be used in functional studies of plant pathogen antagonism. Several novel multifunctional plant growth promoting bacterial strains were earlier isolated from compost (Fui et al., 2017), indicating multifunctional value of compost to plants when applied on the soil.
The authors have not declared any conflict of interests.
The authors are very grateful to Sida/SAREC for the award of scholarship to the first author. Partial research funding from Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS) and from Carl Tryggers Stiftelse (CTS) is gratefully acknowledged.
REFERENCES
|
Acosta-Martínez V, Dowd S, Sun Y, Allen V (2008). Tag-coded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol. Biochem. 40:2762- 2770.
Crossref
|
|
|
|
Buckley DH, Huangyutitham V, Nelson TA, Rumberger A, Thies JE (2006). Diversity of Planctomycetes in soil in relation to soil history and environmental heterogeneity. Appl. Environ. Microbiol. 72:4522-4531.
Crossref
|
|
|
|
|
Chandna P, Nain L, Singh S, Kuhad RM (2013). Assessment of bacterial diversity during composting of agricultural byproducts. BMC Microbiol. 13:99.
Crossref
|
|
|
|
|
Chen MY, Tsay SS, Chen KY, Shi YC, Lin YT, Lin GH (2002). Pseudoxanthomonas taiwanensis sp. nov., a novel thermophilic, N2O-producing species isolated from hot springs. Int. J. Syst. Evol. Microbiol. 52:2155-2161.
Crossref
|
|
|
|
|
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009). The ribosomal database project: improved alignment and new tools for rRNA analysis. Nucleic Acids Res. 37:D141-D145.
Crossref
|
|
|
|
|
Danon M, Franke-Whittle IH, Insam H, Chen Y, Hadar Y (2008). Molecular analysis of bacterial community succession during prolonged compost curing. FEMS Microbiol. Ecol. 65:133-144.
Crossref
|
|
|
|
|
de Bertoldi M, Vallini G, Pera A (1983). The biology of composting: A Review. Waste Manag. Res. 1:157-176.
Crossref
|
|
|
|
|
Fui C, Chin S, Furuya Y, Mohd-Zainudi HM, Ramli N, Hassan MA, Tashiro Y, Sakai K (2017). Novel multifunctional plant growth–promoting bacteria in co-compost of palm oil industry waste. J. Biosci. Bioeng. 5:506-513.
|
|
|
|
|
Fullwood MJ, Wei CL, Liu ET, Ruan Y (2009). Next-generation DNA sequencing of paired-end tags (PET) for transcription and genome analyses. Genome Res. 19:521-532.
Crossref
|
|
|
|
|
Han Z, Liu Y, Zhong M, Shi G, Li Q, Zeng D, Zhang Y, Fei Y, Xie Y (2018). Influencing factors of domestic waste characteristics in rural areas of developing countries. Waste Manag. 72:45-54.
Crossref
|
|
|
|
|
Horisawa S, Sakuma Y, Nakamura Y, Doi S (2008). Profiling of a microbial community under confined conditions in a fed-batch garbage decomposer by denaturing gradient gel electrophoresis. Bioresour. Technol. 99(8):3084-3093.
Crossref
|
|
|
|
|
Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R (2009). A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 3:442-453.
Crossref
|
|
|
|
|
Karadag D, Özkaya B,Ölmez E, Nissilä ME, Çakmakçı M, Yıldız S, Puhakka JA (2013). Profiling of bacterial community in a full-scale aerobic composting plant. Int. Biodeterior. Biodegr. 77:85-90.
Crossref
|
|
|
|
|
Kowalchuk GA, Naoumenko ZS, Derikx PJL, Felske A, Stephen JR, Arkheapchenko IA (1999). Molecular analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in compost and composted materials. Appl. Environ. Microbiol. 65:396-403.
|
|
|
|
|
Lebedinsky AV, Chernyh NA, Bonch-Osmolovkaya EA (2007). Phylogenetic systematics of microorganisms inhabiting thermal environments. Biochemistry (Mosc) 72:1299-1312.
Crossref
|
|
|
|
|
Lipski A, Altendorf K (1997). Identification of heterotrophic bacteria isolated from ammonia-supplied experimental biofilters. Syst. Appl. Microbiol. 20:448- 457.
Crossref
|
|
|
|
|
Liu Z, Lozupone C, Hamady M (2007). Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 35:e120.
Crossref
|
|
|
|
|
MacLean D, Jonathan D, Jones G, Studholme DJ (2009). Application of ´next-generation' sequencing technologies to microbial genetics. Reviews 7:287-296.
|
|
|
|
|
Manyl-Loh CE, Mamphweli SN, Meyer EL, Gakaka G, Simon M, Okoh AL (2016). An overview of the control of bacterial pathogens in cattle manure. Int. J. Environ. Res. Public Health 13:843.
Crossref
|
|
|
|
|
Michel FC, Marsh TJ, Reddy CA (2002). Bacterial community structure during yard trimmings composting. In: Insam, H., Riddech, S., Klammer, S., (Eds.) Microbiology of composting, Springer, Berlin, Heidelberg, New York. pp. 25-43.
Crossref
|
|
|
|
|
Nakagawa Y, Sekane T, Yotota A (1996). Transfer of "Pseudomonas riboflavia" (Foster 1944), a gram-negative, motile rod with a long-chain 3-hydroxy fatty acids, to Devosia riboflavina gen. nov., sp. nov., nom. Rev. Int. J. Syst. Bacteriol. 46:16-22.
Crossref
|
|
|
|
|
Neher DA, Weicht TR, Bates ST, Leff JW, Fierer N (2013). Changes in bacterial and fungal communities across compost recipes, preparation methods, and composting times. PLoS ONE 8(11):e79512.
Crossref
|
|
|
|
|
Ntougias S, Zervakis GI, Ehaliotis C, Kavroulakis N, Papadopoulou KK (2006). Ecophysiology and molecular phylogeny of bacteria isolated from alkaline two-phase olive mill wastes. Res. Microbiol. 157:376-385.
Crossref
|
|
|
|
|
Peters S, Koschinsky S, Schwieger F, Tebbe CC (2000). Succession of microbial communities during hot composting as detected by PCR-Single-Strand-Conformation-Polymorphism-based genetic profiles of small-subunit rRNA genes. Appl. Environ. Microbiol. 66:930-936.
Crossref
|
|
|
|
|
Rebollido R, Martínez J, Aguitera Y, Melchor K, Koerner I, Stegmann R (2008). Microbial populations during composting process of organic fraction of municipal solid waste. Appl. Ecol. Environ. Res. 6:61-67.
Crossref
|
|
|
|
|
Riddech N, Klammer S, Insam H (2002). Characterization of microbial communities during composting of organic wastes. In: Insam H., Riddech, R., Klammer, S., (Eds.) Microbiology of Composting. Springer Verlag, Heidelberg, pp. 43- 52.
Crossref
|
|
|
|
|
Rivas R, Willems A, Subba-Rao NS, Mateos PF, Dazzo FB, Kroppenstedt RM, Martínez-Molina E, Gillis M, Velázquez E (2003). Description of Devosia neptuniae sp. nov. that nodulates and fixes nitrogen in symbiosis with Neptunia natans, an aquatic legume from India. Syst. Appl. Microbiol. 26:47-53.
Crossref
|
|
|
|
|
Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, Daroub SH, Camargo FA, Farmerie WG, Triplett EW (2007). Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283- 290.
Crossref
|
|
|
|
|
Ryckeboer J, Mergaert J, Vaes K, Klammer S, De Clercq D, Coosemans J, Insam H, Swings J (2003). A survey of bacteria and fungi occurring during composting and self-heating processes. Ann. Microbiol. 53:349-410.
|
|
|
|
|
Shendure J, Ji H (2009). Next-generation DNA sequencing. Nat. Biotechnol. 26:1135 -1145.
Crossref
|
|
|
|
|
Sneath PHA (1986). Endosoreforming gram-positive rod and cocci. In: Sneath, P.H.A., Sharpe, M.E., Holt, JG., (Eds.) Bergey's Manual of Systematic Bacteriology 2:1104-1207.
|
|
|
|
|
Strous M, Fuerst JA, Kramer HM, Logemann S, Muyzer G, van de Pas-Schooner KT, Webb R, Kuenen JG, Jetten MS (1999). Missing lithotroph identified as new planctomycete. Nature 400:446- 449.
Crossref
|
|
|
|
|
Song C, Li M, Jia X, Wei Z, Zhao Y, Xi B, Zhu C, Liu D (2014). Comparison of bacterial community structure and dynamics during the thermophilic composting of different types of solid wastes: anaerobic digestion residue, pig manure and chicken manure. Microb. Biotechnol. 7(5):424-433.
Crossref
|
|
|
|
|
Sundberg C, Franke-Whittle HI, Kauppi S, Yu D, Romantschuk M, Insam H, Jönsson H (2011). Characterisation of source-separated household waste intended for composting. Bioresour. Technol. 102:2859-2867.
Crossref
|
|
|
|
|
Székely AJ, Sipos R, Berta B, Vajna B, Hajdú C, Márialigeti K (2008). DGGE and T-RFLP analysis of bacterial succession during mushroom compost production and sequence-aided T-RFLP profile of mature compost. Microb. Ecol. 57:522-533.
Crossref
|
|
|
|
|
Taiwo LB, Oso BA (2004). Influence of composting techniques on microbial succession, temperature and pH in a composting municipal solid waste. Afr. J. Biotechnol. 3:239-243.
|
|
|
|
|
Takaku H, Kodaira S, Kimoto A, Nashimoto M, Takagi M (2006). Microbial communities in the garbage composting with rice hull as an amendment revealed by culture-dependent and independent approaches. J. BoiSci. BioEng. 101:42-50.
Crossref
|
|
|
|
|
Tumuhairwe JB, Tenywa JS, Otabbong E, Stig L (2009). Comparison of four low-technology composting methods for market crop wastes. Waste Manag 29:2274 – 2281.
Crossref
|
|
|
|
|
Ueda K, Ohno M, Yamamoto K, Nara H, Mori Y, Shidama M, Hayashi M, Oida H, Terashima Y, Nagata M, Beppu T (2001). Distribution and diversity of symbiotic thermophiles, Symbiobacterium thermophilum and related bacteria, in natural environments. Appl. Environ. Microbiol. 67:3779-3784.
Crossref
|
|
|
|
|
Vivas A, Moreno B, Garcia-Rodriguez S, Benitez E (2009). Assessing the impact of composting and vermicomposting on bacterial community size and structure, and microbial functional diversity of an olive-mill waste. Bioresour. Technol 100:1319-1326.
Crossref
|
|
|
|
|
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxanomy. Appl. Environ. Microbiol. 73:526-5267.
Crossref
|
|
|
|
|
Xi B, Zhao X, He X, Huang C, Tan W, Gao R, Zhang H, Li D (2016). Successions and diversity of humic-reducing microorganisms and their association with physical-chemical parameters during composting. Bioresour. Technol. 219:204-211.
Crossref
|
|
|
|
|
Zing X, Lie Y, Zhu L, Cui H, Jia L, Xie X, Lie J, Wei Z (2017). Assessing the use of compost from multiple sources based on the characterization of carbon mineralization in soil. Waste Manag. 70:30-36.
Crossref
|
|
|
|
|
Zhao X, He X, Xi B, Gao R, Tan W, Zhang H, Li D (2016). The evolution of water extractable organic matter and its association with microbial community dynamics during municipal solid waste composting. Waste Manag. 56:79-87.
Crossref
|
|