ABSTRACT
This review aimed to make an inventory of the relevant work carried out on antibiotic resistance in the animal sector during the last two decades in French-speaking countries of the West African sub-region. English and French published articles from 2000 to 2019 indexed in PubMed, Google Scholar, and African Journals Online were reviewed in accordance with an adapted PRISMA guideline. Mean Resistance (MR) and interquartile ranges (IQR) of resistance were calculated for each antibiotic-bacterium combination for each country and globally. 28 articles were eligible for this qualitative review. One third of the countries did not have suitable data on antibiotic resistance in animals. Senegal (11/28) and Ivory Coast (8/28) are at the top of countries where more studies have been carried out. Poultry (17/28), cattle (10/28) and pigs (4/28) are the most investigated species. In poultry, resistance in E. coli strains was high to Tetracycline’s (MR: 97%; IQR [80.65%- 98.5%]). Resistance in Salmonella spp. strains from poultry was high to Erythromycin (MR: 100%; IQR [99%-100%] and Amoxicillin-Clavulanic acid (47.76%; IQR [16.06%-52.52%]). In cattle, resistance of Staphylococcus spp. was low in general for all antibiotics with resistance of 16.25% IQR [11.75%-20.58%], 14.63% IQR [13.82%-31.32%], 10% IQR [8.55%-16%] respectively for Tetracycline’s, Penicillin, and Gentamicin. More studies deserve to be done in West Africa French speaking countries in order to draw attention of decision-makers, lead to regulations on the correct use of antibiotics in the veterinary sector, and if possible set up a sub-regional network for the monitoring of antibiotic resistance.
Key words: Antibiotic resistance, animals, West Africa, French countries.
Antimicrobial resistance (AMR) both in human and veterinary medicine has reached alarming levels in most parts of the world and has been recognized as a significant emerging threat to global public health and food security. West African countries face, like the rest of the world, this serious problem of the emergence of resistances to antibiotics (Ouédraogo and Sylvain, 2017). To fight against this threat, joint resolutions and actions are promoted by international organizations to combat AMR globally (Wall et al., 2016). Antimicrobial resistance affecting humans and animals is primarily influenced by an increase in using antimicrobials for a variety of purposes, including therapeutic and non-therapeutic uses in animal production. These practices contribute to the spread of drug-resistant pathogens in both livestock and humans (Van Boeckel et al., 2015). In low- and middle-income countries including African sub-Saharan countries, population growth and rising incomes have driven an unprecedented growth in demand for animal protein. As a result, efforts to meet rising demand are driving a shift in animal production from small holder, mixed crop, and livestock operations to increasingly intensive, large-scale, and specialized commercialization farms (Schar et al., 2018). These intensive production systems are known to employ more and more antimicrobial for therapeutic and non-nontherapeutic use, including mass administration for prevention and control of disease and as growth promoter. Unfortunately, in most African sub-Saharan countries, it is known that there is no or less control over the distribution of veterinary pharmaceuticals and phytosanitary products. Worse still, no appropriate legislation yet exists to guarantee the quality of the various antimicrobial products released onto the market. In West African French Speaking countries for example, there are massive shortcomings in the organization of the veterinary drug market. These include lack of specific legislation following the recent liberalization of veterinary drugs, lack of veterinary drug inspections before marketing drugs, lack of registration because of the existence of parallel channels alongside the official distribution channel of veterinary drugs (Mensah et al., 2014). The consequence of all these shortcomings is, in one side, the large scale of antimicrobial misuse in farms to combat low productivity and high mortality caused by infectious diseases and, in the other side, the development of resistances to antimicrobials. Unfortunately, there is limited data concerning anti-microbial use and antimicrobial resistance in these countries in comparison with English speaking countries of the West African Sub region due to the absence of systematic surveillance systems. Thus, conclusions must be drawn from point-prevalence assessments or research studies. Here, the available information are piece together to build a picture of the situation of resistance development in food producing animals in West African French speaking countries. Food producing animals are linked to humans via the food chain and shared environment (Oloso et al., 2018). Thus, they can play an important role in the dissemination of resistance pathogens or resistance genes to humans (Chantziaras et al., 2014). This review over his importance in animal health will have an importance in public health because it offers the opportunity to see the burden of Antibiotic resistance in the region.
Research questions and objective
Some research questions guide this study, which aimed to establish the situation of ABR in food producing animals in 09 West African French speaking countries. These questions were: (i) what is the status of antibiotic resistance (ABR) in the food producing animals according to previous studies in the two last decades? (ii) what is the pattern of resistance in each state? (iii) what is the status of ABR among the common antibiotics that are used to control pathogens at animal’s level?
Data search design
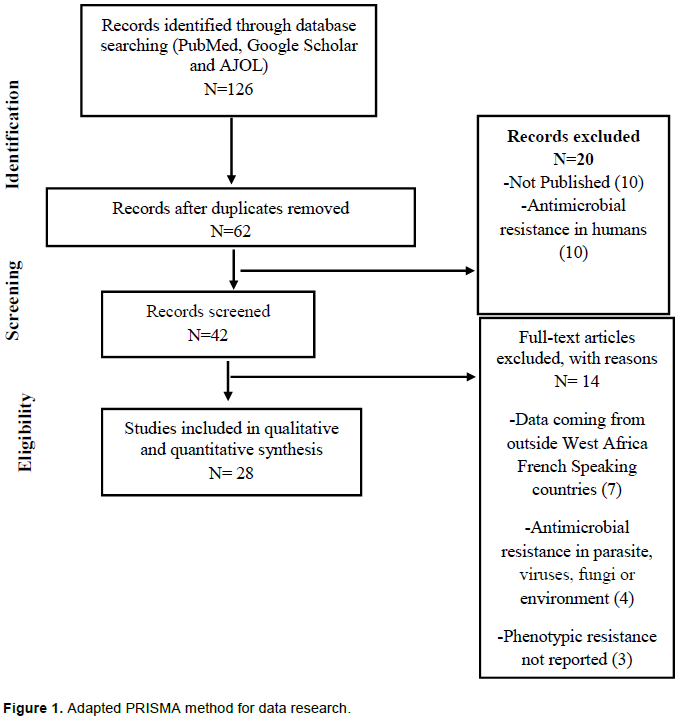
Free databases (Pub Med, Google Scholar and African Journals Online) were searched using broad terms in English and French, “antimicrobial, resistance, and country name”. Where necessary, search terms were stated as strings: Antimicrobial resistance OR Antimicrobial susceptibility AND country name AND animals; “animals” was substituted with different animal names (poultry, goat, sheep, cattle, camel, pig, etc.). References in the identified materials were also searched. Indeed, the reference lists of all included articles were used to carry out a supplementary literature search. Review articles in English were retrieved and assessed for potential relevant studies related to ABR in food producing animals in West Africa French speaking countries. The PRISMA-style flowchart was modified and used for this review (Figure1). Each publication was treated as a study, which contains single or multiple reports.
Exclusion and inclusion criteria
Articles were assessed using predesigned eligibility forms and according to predefined eligibility criteria (Table 1). Briefly, studies on parasites, viruses, and fungi were excluded. Studies dealing with ABR in humans were excluded. Studies reporting data from outside West Africa French speaking countries were not further selected. The selection of French and English published articles was based on clearly defined populations involving food animals at farms and/or processed/freshly slaughtered animals at abattoirs/markets. To be included, studies must have performed antibiotic susceptibility testing with antibiotics using appropriate methods and results interpreted according to appropriate guidelines.
Literature screening and data extraction
Mendeleyev (version 1.19.4) was used for literature management, and relevant data from included articles were extracted. The data were abstracted and analyzed using a framework on an Excel (Microsoft Office Excel 2013) spreadsheet. Each study included Country, first author details, year of publication, aims, study population (such as, pigs, poultry, cattle, sheep, goat), type of sample (such as, nasal swabs, rectal swabs, fecal samples, and meat products), sample size, clinical status (such as, apparently healthy, sick, and dead), study site (slaughterhouse, farm, and market), type of study (cross sectional, longitudinal), bacteria of interest (such as, Staphylococcus aureus, Salmonella spp., Campylobacter spp., Escherichia coli, and Enterococcus spp.), antibiotics tested, antimicrobial susceptibility testing (AST) methods (disk diffusion, micro-broth dilution, agar dilution, E-test, and automated methods), guidelines of interpretation of AST (such as, CA-SFM, EUCAST, CLSI, and NCCLS), ABR prevalence and molecular investigations.

Quality assessment
Preexisting scales were not used to assess study quality. Appraisal tool was used to assess the quality of included studies. So the researcher checked if:
(1) the basic data including: sample type, bacteria of interest, and study site, was provided; (2) samples of the study were collected in an appropriate way; (3) sample size was representative of the target population; (4) the number of strains representative; (5) the Antimicrobial Susceptibility Test perform with a valid method and interpreted according to a valid guideline; (6) all important sub-groups (Animal population or sample) identified and accounted for when reporting resistance rate. If all the criteria were met, the study was ranged according to the quality. If one or more criteria were not fulfilled, the study was ranged as moderate quality.
Data analysis
Microsoft Excel (2013 for Windows) was used to analyze the data following an initial extraction. Prevalence was calculated, median resistance (MR) and interquartile range (IQR) of resistance for each bacterium-antibiotic combination in each specific animal population (poultry, cattle…). Meta-analysis was not conducted because of the small number of articles available. All reports reporting ABR for an antibiotic were categories as no resistance (when resistance was <1%); very low resistance (1-24%); low resistance (25-49%); high resistance (50-74%) and very high resistance (75-100%) (Figure 1).
Number of studies per country and per year
A total of 28 studies from 6 countries were included in the review (Vounba et al., 2018, 2019; Cardinale et al., 2003; Bada-Alambedji et al., 2006; Fall-Niang et al., 2019; Fall et al., 2012; Shyaka et al., 2010; Stevens et al., 2006; Dione, 2009; Kadja et al, 2013; Mama et al., 2019; Sidibé et al., 2019; Coulibaly et al., 2010; Rene et al., 2014; Attien et al., 2013; Gblossi et al., 2012; Abdoulkarim et al., 2013, 2014; Caroline et al., 2019; Kagambèga et al., 2013; Somda et al., 2018; Deguenon et al., 2019; Boko et al., 2013; Ahouandjinou et al., 2016; Yao et al., 2018, 2017; Coulibaly et al., 2018; Guessennd et al., 2012. Three out of nine countries had no suitable report on antimicrobial resistance in animals as shown in Figure 2. The country in the considered region having the higher number of reports is Senegal followed by Ivory Coast. The majority of published articles are from the last decade as shown in Figure 3. The mean of published papers per year is less than 2 with 2019 being the year with a record number of published papers.
Situation of ABR in food producing animals in all countries
Animal population and bacteria of interest in studies
Investigations were mainly done in poultry (60.71%; 17/28) and cattle (35.71; 10/28) followed by Pigs (14.28%; 4/28) Sheep’s (7.14%; 2/28), Goats (3.57%; 1/28) and Guinea fowl (3.57%; 1/28). Some studies investigated two or more species at the same time. The investigated bacteria were Salmonella spp. (50%; 14/28); Staphylococcus spp. (25%; 7/28); Escherichia coli (10.7; 3/28) and Campylobacter spp. (7.14%; 2/28). These bacteria were tested against 54 antibiotics belonging to 14 classes (Table 2). The most tested ATB were Tetracyclines (23 reports) followed by Gentamicin and association Sulfoxide-Trimethoprim (22 reports each). High to very high resistances were frequently reported for Tetracyclines (8 reports) followed by Sulfoxide-Trimethoprim, Streptomycin and Amoxicillin-clavulanic acid with 05 reports each (Table 3).
Samples tested and their origin
Studies included in this review investigated mainly in healthy animals (75%). Studies in diseased animals were performed in animals with mastitis (14.29%), Colibacillosis (7.14%) or Salmonellosis (7.14%). The majority of the studies were performed in samples collected in slaughterhouses or vendors in markets (60.71%; 17/28) and farms (46.42%; 13/28). Only one study was undertaken in samples from veterinary clinic and three studies investigated samples from both farm and slaughterhouse. Samples investigated were mainly carcasses (39.29% of the studies) and feces (39.29%). Two studies investigated carcass and feces at the same time. Milk was investigated in 4 studies (14.28%) and only two studies (7%) investigated animal organs. Nasal and cloacal swabs were used as samples in one study each. Table 4 resumes the characteristics of included studies.
Resistance in poultry
Resistance was analyzed in E. coli, Staphylococcus spp. And Salmonella spp. subgroup. Resistance in E. coli strains from poultry was high to Tetracycline (MR: 97%; IQR [80.65- 98.5%]); Sulfoxide (MR: 81.4%; IQR [81.1-81.7%]); Sulfoxide-Trimethoprim (MR: 61%; IQR [46.57-68.85]) and Ampicillin (42.68%; IQR [40.58-60.43%]). Reported resistance in Salmonella strains from poultry was high to: Erythromycin (MR: 100%; IQR [99%-100%]; Amoxicillin-Clavulanic acid (47.76%; IQR [16.06-52.52%]); and Tetracyclines (46.04%; IQR [38.06-60.75]). Resistance of Campylobacter spp. to ciprofloxacin was 60% (IQR [55.25-65.75%])
Resistance in cattle
E. coli strains isolated from cattle’s show very high resistances with resistance of 98.8% (IQR [98.2-99.4%]) for Tetracycline’s; 97.65% (IQR [96.48-98.43%]) for Aztreonam and 96.45% (IQR [94.68-98.23]) for Ampicillin. Resistance of Salmonella spp. strains to Streptomycin was high, 58.58% (IQR [40.04-77.11%]), and resistance of Staphylococcus spp. was low in general for all antibiotics with resistance of 16.25% IQR [11.75-20.58%], 14.63% [13.82-31.32%], 10% [8.55-16%] respectively for Tetracycline’s, Penicillin and Gentamicin.
Molecular investigations
Resistance and virulence genes in E. coli
Three studies investigated AMR and or virulence genes in E. coli strains. Somda et al. (2018) in BF investigated the presence of STEC, EPEC, ETEC, EIEC, and EAEC genes on grilled chicken meat samples by 16- plex PCR for the genes uidA, pic, bfp, invE, hlyA, elt, ent, escV, eaeA, ipaH, aggR, stx1, stx2, estIa, estIb, and ast. Only stIa, stx2A, invE, astA, and aggR virulence genes were detected. Six diarrheagenic E. coli were detected as follows: EAEC and ETEC (in two samples each) and STEC and EIEC (in one sample each). No EPEC gene was detected.
In Senegal, Vounba et al. (2018) investigated Antimicrobial Resistance (AMR) genes in E. coli isolated from diseased chicken. Many AMR genes were detected, including variants of blaCTX-M encoding resistance to third-generation cephalosporins. Most fluoroquinolone-nonsusceptible isolates were carriers of mutations in gyrA (Ser83Leu, Asp87Asn, and/or Asp87Tyr) and/or parC (Ser80Ile) genes. A total of 84.5% isolates exhibited at least one of the virulence markers of Avian Pathogenic Escherichia Coli (APEC), among which 39.7% were defined as potential virulent APEC. The same author investigated E. coli from healthy chicken in Senegal (Vounba et al., 2019) and reported the presence of AMR genes. According to this report, 95% of tested farms harbored isolates carrying mutations in gyrA (Ser83Ile and Asp87Asn) and parC (Ser80Ile). 3GC resistance was mediated by blaCMY-2 or blaCTX-M genes, blaCTX-M bein Stevens g of genotypes blaCTX-M-1, blaCTX-M-8 and the genotype blaCTX-M-15. The most prevalent AMR genes were those encoding resistance against tetracycline (tet genes) and trimethoprim-sulfamethoxazole (dfrA genes). Genes encoding resistance against streptomycin (aadA1) or ciprofloxacine (qnrB) were also detected.
Resistance and virulence genes in Salmonella strains
Boko et al. (2013) screened all confirmed Salmonella isolates in Guinea fowl by polymerase chain reaction (PCR) for the presence of several virulence-associated genes located on Salmonella Pathogenicity Islands (SPI) 1 to 5: prgH, invA, sitC, spaI, invE (SPI-I); spiC, ssaU, ttrB (SPI-2); mgcT, misL (SPI-3); orfL (SPI-4); pipD (SPI-5). All isolates belonging to five serotypes tested positive by PCR for most of the target genes (Deguenon et al., 2019). The genes of virulence that were targeted for amplification by PCR were invA, spvR, spvC, fimA and stn. All Salmonella isolates were positive for the presence of invA genes, fimA and stn. The spvC gene was present in 10% and spvR gene in 20% of the isolates.
Resistance and virulence genes in Staphylococcus strains
Mama et al., 2019 in Senegal tested six S. aureus isolates of cow origin. All the isolates showed resistance for penicillin (with blaZ gene) and two of them to tetracycline (with tet (K) gene). All the isolates hosted hemolysin-encoding genes. The following genotypes were observed for non S. aureus strains: tetracycline [tet(K), tet(L)], trimethoprim/sulfamethoxazole (SXT) (dfrG, dfrK), penicillin (blaZ), erythromycin [msr(A)/ msr(B)].
In Niger, Abdoulkarim et al., 2014 tested all the isolates resistant to Amoxicillin in his study for the presence of blaZ gene (coding for β-lactamase) by polymerase chain reaction (PCR). 90% of the isolates tested positive for the blaZ gene suggesting that the blaZ gene encodes the production of β-lactamase by most penicillin-resistant S. aureus of this study. Previously, the same author (Abdoulkarim et al., 2013) virulotyped S. aureus strains by PCR for the presence of genes coding for Microbial Surface Components Recognizing Adhesive Matrix Molecules (MSCRAMM) to colonize the host epithelia and extracellular matrix components (clfA, clfB, fnbA, cna, ebpS and sdrC). This was done for capsular and other surface antigens conferring resistance to phagocytosis (cap5H, cap8H, spa and icaA); exfoliative toxins active on extracellular matrix components (etA, etB and etD); haemolysins and leukocidins counteracting the cells of the host’s innate immune response (hla, hlb, hld, hlgAC, lukD, lukM, lukF-PV and lukS-PV); and for enterotoxins causing diarrhoea by action on the host enterocytes (sea, seb, sec, sed, seg, seh, sei, sej and sen). All or most of the S. aureus isolates gave amplified fragments with most for the genes coding for surface antigens: clfA (91%), clfB (100%), fnbA (100%), cna (100%), ebpS (100%), sdrC (87%), cap5H (83%), spa (96%), icaA (96%); and with several for the toxin-encoding genes: etD (74%), hla (96%), hlb (100%), hld (96%), hlgAC (74%), lukD (96%), lukM (96%). Conversely, a minority of isolates tested positive for the following genes: cap8H (4%), lukF-PV (17%), lukS-PV (9%); only one isolate (4%) tested positive for 2 of the 9 enterotoxin-encoding genes (sej and seg);
Situation of ABR per country
Senegal
Eleven studies from Senegal are included in this qualitative review. 39 different antibiotics were tested in these studies namely : Methicillin ; Penicillin; Ampicillin; Amoxicillin; Amoxicillin+ Clavulanic Acid; Nalidixic Acid; Cefotaxime; Ceftazidime; Ticarcilline; Imipenem; Cefoxitine; Cephalexin; Cephoperazone; Cefalotin; Cefoxitin; Ceftiofur; Ceftriaxone; Flumequine; Gentamicin; Spectinomycine; Streptomycin; Spiramycine; Kanamycin; Amikacin; Pefloxacin; Norfloxacin; Ciprofloxacin; Neomycin; Erythromycin; Tetracycline; Doxycycline; Chloramphenicol; Nitrofurane; Sulfoxide; Sulfoxide-Trimethoprim; Tobramycin; Trimethoprim; Colistin; Rifampicin; Tetracyclines were the most frequently tested antibiotics in the studies (90.9%) followed by the association Sulfoxide-Trimethoprim (81.81) and Gentamicin tested in 72.72% of the studies.
Most of the studies in Senegal were performed in poultry (63.63%; 7/11). Four studies out of the 7 in poultry was performed in carcass sampled in slaughterhouses or vendors and 2 studies performed on feces sampled in farms, one (1/7) on animal’s organs and one on nasal samples. Salmonella (3/7), E. coli (2/7), Campylobacter (1/7) and Staphylococcus spp. (1/7) were the studied bacteria in poultry. The ABR in Campylobacter was 75% to Ciprofloxacin; in Staphylococcus spp., resistance was 83.33% for Tetracycline and 50% for Sulfoxide-Trimethoprim.
In Salmonella spp. the resistance was mainly observed to Tetracyclines (66%; [56.34-70.5]), Trimethoprim (59.1%; [50.55%-67.65%); sulfoxides (56.55%; [48.83-64.28%]) Streptomycin (50.86%, IQR: [39.33-62.44]) and association Sulfoxide-Trimethoprim (47% [43.5-63.5]). The highest resistance in E. coli was observed for the same antibiotics: Tetracycline (98.50%; [97.75-99.25%]), Sulfoxide (81.04% [81.1-81.7%]) Sulfoxide-Trimethoprim (68.85%; [64.93-72.78]) and Ampicillin (58.15%; [48,23%-68,08%]). Two studies focus on cattle (2/11; 18.18%) and bacteria of interest was Staphylococcus spp. (milk samples) for one study and Salmonella spp. (carcass samples) for the other study. Only one study on ABR was performed in pigs (Carcass and feces samples) and one on small ruminants (Sheep and goat with milk as sample matrix). Staphylococcus spp. as bacteria of interest was investigated in these two studies.
Ivory coast
Twenty eight different antibiotics were tested in a total of 8 studies included in this qualitative review: Pristinamycin; Oxacillin; Ofloxacin; Amoxicillin; Amoxicillin+Clavulinic Acid; Nalidixic Acid; Cefotaxime; Cefuroxime; Imipenem; Cefalotin; Ceftriaxone; Gentamicin; Kanamycin; Rifampicin; Vancomycin; Lyncomycine; pefloxacin; Ciprofloxacin; Erythromycin; Tetracycline; Doxycycline; Chloramphenicol; Sulfoxide-Trimethoprim; Sixomicin; Tobramycin; Piperacillin; Cefepime; Minocycline. The association Amoxicillin-Acid Clavulanic was tested in all the studies followed by Association Sulfoxide-Trimethoprim tested in seven of the eight studies.
Studies were mainly conducted in poultry (4/8) and cattle (3/8) and Salmonella was the bacteria of interest in four studies followed by E. coli in three studies. Globally, the highest prevalence of resistance in Salmonella in poultry was observed with Amoxicillin (50.47% IQR [49.55-51.37]); Amoxicillin-Acid Clavulanic (MR=47.76% [44.50-50,01]) and Cefalotin (MR=42.99%; IQR[41.77%-44.22%]). In cattle, Salmonella was most frequently resistant to Tetracycline’s (MR=42.99%; IQR [41.77%-44.22%]). One of the three studies in poultry investigated resistance of Campylobacter spp. to antibiotics and reported resistances to these bacteria. Indeed, interesting resistances were reported to: Nalidixic Acid (78%) Ciprofloxacin (50%) ; Erythromycin (13,5%) and Amoxicillin + Clavulanic Acid (11.8%) (Gblossi et al., 2012).
Another study investigated the resistance of Staphylococcus spp. in three species (beef pork and chicken) but did not report resistance per specie (Attien et al., 2013). According to this study, resistance to staphylococcus in food producing animals in Ivory Coast was 100% to erythromycin, 62% to Amoxicillin-acid-clavulanic and 58% to Oxacillin. To finish, Guessennd et al. (2012)investigated E.coli resistance to antibiotics in pigs and reported notable resistance of 76.7, 66.7 and 56.7% respectively for Tetracycline, Cefotaxime and Sulfoxide-Trimethoprim.
Benin
Three studies were identified from Benin and antibiotic susceptibility was performed against 24 antibiotics namely: Oxacillin; Ampicillin; Amoxicillin; Amoxicillin-Clavulanic; Nalidixic Acid; Cefotaxime; Cefuroxime; Imipenem; Cefoxitine; Cefalotin; Ceftriaxone; Flumequine; Gentamycin; Amikacin; Ciprofloxacin; Neomycin; Tetracycline; Chloramphenicol; fosfomycine; Sulfoxide; Sulfoxide-Trimethoprim; Trimethoprim; Colistin; gentamicin.
Studies in Benin were done on poultry/sheep/pigs for one study, guinea fowl for another study and cattle. Two studies were performed in feces samples and one on carcass from cattle. Salmonella spp. strains were investigated in the three studies. Salmonella isolates from poultry/sheep/pigs were resistant to Amoxicillin (100%), Amoxicillin-Acid clavulanic (100%) Cefotaxime (100%) Cefoxitin (100%) Cefalotin (100%) Gentamycin (100%) Amikacin (100%) Trimethoprim (100%); Ceftriaxone (85%). Those from Guinea flow were resistant to Oxacillin (100%), Sulfoxide (100%) and Colistin (100%). In cattle, High resistance was reported from Ampicillin (87.77), Ceftriaxone (88.49), Sulfoxide-Trimethoprim.
Burkina Faso
In BF, three published studies reported ABR in food producing animals. The three studies tested a total of 21 antibiotics: Ampicillin; Amoxicillin-Clavulanic acid; Nalidixic Acid; Aztreonam; Cefotaxime; Mecilinam; Ticarcilline; Imipenem; Cephalexin; Ceftriaxone; Gentamycin; Streptomycin; Ciprofloxacin; Norfloxacin; Erythromycin; Tetracycline; Chloramphenicol; Sulfoxide; Sulfoxide-Trimethoprim; Trimethoprim; Colistin.
Common antibiotics in the studies were: Imipenem; Gentamycin; Streptomycin; Ciprofloxacin; Tetracycline and Chloramphenicol. Disc diffusion method was used in all studies. Salmonella and E. coli were the bacteria studied. Salmonella resistance was reported in two studies and E. coli in One study. Resistance of E. coli to Tetracycline in chicken meat was 64.3%. Salmonella from food producing animals in BF was resistant to Erythromycin (MR=100%); Amoxicillin-Clavulanic (MR=55.85%; [54.23-57.48%]); Ticarcillin (MR=49%; [45.78-53.13%]) and Tetracycline (MR=42.1%; IQR [34.05-43.75%]).
Mali-Niger
Three published studies were found for these countries. In Mali, the only published paper reported resistance to salmonella in poultry while in Niger the two included studies reported resistance of Staphylococcus spp. in Cattle milk. The three studies used disc diffusion method and followed the CASFM guidelines. Resistance of salmonella in Mali was high for Erythromycin (98%); Colistin (94%); Streptomycin (90%); Kanamycin (67%); Flumequine (65%) and Tetracycline (59%). In Niger, moderate to low resistance of S. aureus was observed to penicillin (30.5% [21.75-39.25]); Gentamycin (16% [13-19]); and Tetracycline (10.5% [9.25-11.75]).
As a threat to a century of gains made since the discovery of antibiotics and the contribution of these drugs to improvements in animal and human health and wellbeing, antibiotic resistance has become a global concern. Sub-Saharan Africa has a high incidence of invasive bacterial infections in humans and this condition increase demand for both preventive and therapeutic antimicrobials. However, antibiotics are widely used in domestic and commercial animal husbandry and this can contribute to the annihilation of the therapeutic arsenal used in human medicine and on which the continent's and the world relies to overcome these infections. Indeed, the development of resistances in animals can threat human health through dissemination of resistance bacteria or resistance genes from animal to human through food chain or direct contact.
Being in human health or veterinary medicine, sub-Saharan Africa has the least antimicrobial surveillance strategies of all world regions. Only six (15%) of the 41 WHO Africa region member states carry out surveillance for bacterial antimicrobial resistance in human health (Williams et al., 2018). In animal health, surveillance of antibacterial resistance in animals is almost nonexistent in Africa countries. Then, the lack of consistency in the measurement and reporting of antibiotic susceptibility data in animals makes it difficult to know the situation in food producing animals in different countries. To address this issue, scientists are making efforts in research to yield data that can draw attention of decision makers to set up surveillance network. Unfortunately, these efforts although useful are often rare and scattered that they do not allow the visibility of the magnitude of the situation. That is why a synthesis of the scientific data generated here and there over a long period is often useful to give visibility of the situation and draw attention. This summary of the situation of antibiotic resistance in food producing animal in West Africa French-speaking countries from 2000 to 2019 (20 years) trailed this objective.
In a recent systematic review and meta-analysis, some published papers tried to report the situation of ABR in food producing animals in all African countries (Founou et al., 2018). However, it failed sometime to include some interesting studies from French countries in west Africa may be because some are published in French or are not index in high databases as PubMed or simply did not met authors inclusion criteria. The present review has the advantage of zooming on the situation of antibiotic resistance in French-speaking countries, where interesting scientific work is often not very visible because of language limitations. Indeed, this study proved that antibiotic-resistant is globally under investigation in West Africa French speaking countries with only 28 studies included from 6 countries out of the 9 countries in the study area. This is similar to the situation previously reported by Founou et al. (2018) who reported data from 12 countries out of 54 in Africa. However, unlike this study, which includes only one study from Senegal, many data were reported from Senegal and data from other French speaking countries, giving the real situation in this geographic area.
Our systematic review has demonstrated widespread prevalence of antibiotic resistance in food producing animals in West Africa French speaking countries. Poultry and cattle were the most studied species with Salmonella spp. and Staphylococcus spp. as bacteria’s of interest in most investigations. The fact that poultry is the specie most concerned by studies is not surprising and is rather interesting because poultry farming is the sector that uses antibiotics the most, often in modern farms constituted by semi-intensive or intensive systems unlike other livestock sectors which for the most part still extensive (Schneider et al., 2010). Salmonella as bacteria of interest in studies would be linked to the role of salmonellosis in food poisoning and to the interest of this bacterium in several monitoring programs (Jajere, 2019).
Many studies investigated ABR on samples from healthy animals and this is a good indicator according to WHO advisory group on integrative surveillance who estimated that samples from both healthy animals and sick animals are useful for surveillance but samples from healthy animals should be the primary focus for surveillance because such samples can provide an unbiased measure of antimicrobial resistance in animals source of human food supply (WHO, 2017).
Studied bacteria were Salmonella, Staphylococcus spp., E. coli and Campylobacter spp. . These bacteria are important bacteria commonly investigated and monitored for resistance in many monitoring systems. According to WHO advisory group worldwide, Salmonella is the first priority for inclusion in a program of integrated surveillance of antimicrobial resistance in foodborne bacteria. Campylobacter spp. is also an important foodborne pathogen and should be included in program of integrated surveillance of antimicrobial resistance in foodborne bacteria. Because Escherichia coli are common and some strain variants may cause disease, E. coli is used as a sentinel organism for antimicrobial resistance. E. coli also serve as reservoirs of resistance genes that can be transferred to human pathogens transiting the intestinal tract; as such, it provides information on the flow of Gram-negative resistance trend in the food chain (WHO, 2017).
The majority of studies included used the disk diffusion method and CASFM guidelines. This reduces the impact of the variation in AMR methodology on the pooled estimation. Resistance was found to be high in E. coli strains from poultry to Tetracycline (MR: 97%; IQR [80.65-98.5%]); Sulfoxide (MR: 81.4%; IQR [81.1-81.7%]); Sulfoxide-Trimethoprim (MR: 61%; IQR [46.57-68.85]) and Ampicillin (42.68%; IQR [40.58-60.43%]). Similar high rates of resistance in E.coli have been reported for tetracycline (10.6- 95%), ampicillin (6.02- 95.7%) and trimethoprim/sulfamethoxazole (4.49-80%) in a previous review (Alonso et al., 2016).
In Salmonella strains, high resistance were found to: Erythromycin (MR: 100%; IQR [99-100%]; Amoxicillin-Clavulanic (47.76%; IQR [16.06-52.52%]); and tetracycline (46.04%; IQR [38.06-60.75]). Full resistance to Erythromycin (100%) is not surprising as Salmonella is known to have natural resistance to this antibiotic (Braoudaki and Hilton, 2005). The findings are comparable with previous review in the African region by Founou et al. (2018) who identified similar resistance of Salmonella. Indeed, these authors’ reported a high level of resistance of Salmonella (80.9% [95% CIs; 54-93.8%) in a Meta-analysis of all reported resistance to all antibiotics. This findings highlights serious concerns relating to the use of these antibiotics in animals and corroborate similar findings in Cameroun where Moctar et al. (2019) reported high levels of resistance of E. coli and Salmonella spp. to all classes of antibiotics tested who are usually antibiotics of critical importance for humans and animals according to WHO and OIE classifications. Very few publications associated molecular investigation of resistance to phenotypic resistance. This is because the most important in these countries remain the inventory of the situation of antibiotic resistance before any thorough investigation.
Given the findings of this review, harmonization efforts are urgently needed in West Africa French speaking countries. Standardizing bacteria of interest, antibiotic to be tested (to avoid testing bacteria against antibiotics for which they have a natural resistance such as salmonella against erythromycin which has been reported a lot in studies when this is not really of interest to public health), AMR methods, interpretation guidelines, report format and prioritizing animal population of interest, could allow better comparability of results and situation from all countries. Also in this sense, technical and financial support of laboratories in these countries is a necessity to support the monitoring of the situation in human and food producing animals in a one health approach as recommended by WHO (2017) so as not to undermine the efforts undertaken around the world to combat resistances to antibiotics. The limitations of the current review include the exclusion of English language countries as Nigeria and Ghana biasing this view of the situation in the wall West Africa Sub region. For example, only in Nigeria, with similar inclusion and exclusion criteria, the review of (Oloso et al., 2018)reported 77 studies from animal sector. This shows that the inclusion of the situation in English-speaking countries would provide a better view of the situation in West Africa. A further limitation is combining AMR results from different species across different countries to have median resistance for a bacterium. However, given the observed trends, it is believed that the resolution of the obtained data was sufficient to show general trend of AMR in livestock in French Speaking countries. Moreover, since no monitoring system exist, this review can draw attention of sub regional institutions as ECOWAS to further support the establishment of One Health monitoring networks for the wall countries as it is done in many part of the word especially in Europe with EUCAST monitoring network (Silley et al., 2011).
Tackling the public health threat posed by antimicrobial resistance requires effective antimicrobial resistance surveillance programs. The essential need for robust antimicrobial resistance surveillance systems is emphasized in the Global Action Plan on Antimicrobial Resistance (WHO, 2017). Based on this study, antibiotic resistance is high in food producing animals in West Africa French speaking countries. It is necessary to design a carefully planned, multi-sectoral, surveillance plan, which can be used for research and monitoring of resistances in all countries and sectors in a one health approach. The relevant ministries and governments should enforce registration and monitoring of veterinary drugs use, promote good practices in antimicrobial use by trained professionals.
The authors have not declared any conflict of interests.
REFERENCES
|
Abdoulkarim II, Bada-Alambedji R, Dupreza J, Djikac M, Moulad N, Otea I, Bardiaua M, Mainila JG (2013). Bacterial mastitis in the Azawak zebu breed at the Sahelian experimental station in Toukounous (Niger): Identification and typing of Staphylococcus aureus. International Research Journal of Microbiology 4(7):168-178.
|
|
|
|
Abdoulkarim II, Duprez J, Bada-Alambedji R, Moula N, Mainila JG, Bardiau M (2014). Antibiotic resistance trend of Staphylococcus aureus isolated between 2010 and 2012 from mastitis cases in Azawak zebu in Niger. African Journal of Microbiology Research 8(35): 3271-3275.
Crossref
|
|
|
|
|
Ahouandjinou H, Baba-Moussa F, Bertin G, Sina H, Adéoti K, Mousse W, Pouadjeu-Wouansi S, Toukourou F, Soumanou M, Baba-Moussa L (2016). Antibiorésistance Et Facteurs De Virulence Des Souches D'escherichia Coli Isolées Des Carcasses Bovines Du Bénin. European Scientific Journal 12(33):1857-7881.
Crossref
|
|
|
|
|
Alonso CA, Zarazaga M, Sallem RB, Jouini A, Slama KB, Torres C (2016). Antibiotic resistance in Escherichia coli in husbandry animals: the African perspective. Letters in Applied Microbiology 64:318-334.
Crossref
|
|
|
|
|
Attien P, Sina H, Moussaoui W, Dadieacute T, Chabi SK, Djeacute ni T, Bankole H, Kotchoni S, Edoh V, Preacute vost G, Djegrave M, Baba Moussa L (2013). Prevalence and antibiotic resistance of Staphylococcus strains isolated from meat products sold in Abidjan streets (Ivory Coast). African Journal of Microbiology Research 7(26):3285-3293.
Crossref
|
|
|
|
|
Bada-Alambedji R, Fofana A, Seydi M, Ayayi AJ (2006). Antimicrobial resistance of salmonella isolated from poultry carcasses in Dakar (Senegal). Brazilian Journal of Microbiology 37(4):510-515.
Crossref
|
|
|
|
|
Boko CK, Kpodekon TM, Duprez JN, Imberechts H, Taminiau B, Bertrand S, Mainil JG, Kadoéito Boko C (2013). Identification and typing of Salmonella enterica serotypes isolated from guinea fowl (Numida meleagris) farms in Benin during four laying seasons (2007 to 2010) Identification and typing of Salmonella enterica serotypes isolated from guinea fowl. Avian Pathology 42(1):1-8.
Crossref
|
|
|
|
|
Braoudaki M, Hilton AC (2005). Mechanisms of resistance in Salmonella enterica adapted to erythromycin, benzalkonium chloride and triclosan. International Journal of Antimicrobial Agents 25(1):31-37.
Crossref
|
|
|
|
|
Cardinale E, Dromigny JA, Tall F, Ndiaye M, Konte M, Perrier-Gros-Claude JD (2003). Fluoroquinolone Susceptibility of Campylobacter Strains, Senegal. Emerging Infectious Diseases 9(11):1479-1481.
Crossref
|
|
|
|
|
Caroline BS, Kagambèga A, Bonifait L, Bako E, Cisse H, Bawa I, Le Gall F, Wereme-n'diaye A, Traore S, Chemaly M, Salvat G, Barro N (2019). Serotypes and Multiresistant Salmonella sp. from Chicken Eggs and Laying Hens in Burkina Faso. International Journal of Sciences 8(12):19-25.
Crossref
|
|
|
|
|
Chantziaras I, Boyen F, Callens B, Dewulf J (2014). Correlation between veterinary antimicrobial use and antimicrobial resistance in food-producing animals: a report on seven countries. Journal of Antimicrobial Chemotherapy 69(3):827-834.
Crossref
|
|
|
|
|
Coulibaly EK, Bakayoko S, Karou TG, Coulibaly KJ, Goualie GB, Dosso M, Diopoh KJ (2010). Sérotypage et antibiorésistance des souches de Salmonella isolées dans les foies de poulets vendus sur les marchés de Yopougon (Abidjan Côte d'Ivoire) en 2005. Revue Africaine de Santé et de Productions Animales 8(S):25-29.
|
|
|
|
|
Coulibaly I, Julien CK, Eric-parfait KK, Fatoumata C (2018). Study Of The Antibiotic Resistance Profile Of Escherichia Coli And Salmonella Spp . Isolated From Cattle Dung At The Port-Bouët Slaughterhouse (Abidjan, Ivory Coast). International Journal of Advanced Research and Publications 2(10):5-12.
|
|
|
|
|
Deguenon E, Dougnon V, Lozes E, Maman N, Agbankpe J, Abdel-Massih RM, Djegui, F, Baba-Moussa L, Dougnon J (2019). Resistance and virulence determinants of faecal Salmonella spp. isolated from slaughter animals in Benin. BMC Research Notes 12(317):1-7.
Crossref
|
|
|
|
|
Dione M, Ieven P, Garin B, Marcotty T, Geerts S (2009). Prevalence and antimicrobial resistance of salmonella isolated from broiler farms, chicken carcasses, and street-Vended restaurants in Casamance, Senegal. Journal of Food Protection 72(11):2423-2427.
Crossref
|
|
|
|
|
Fall-Niang NK, Sambe-Ba B, Seck A, Deme SN, Wane AA, Bercion R, Alambedji-Bada R, Gassama-Sow A (2019). Antimicrobial Resistance Profile of Salmonella Isolates in Chicken Carcasses in Dakar, Senegal. Foodborne Pathogens and Disease 16(2):130-136.
Crossref
|
|
|
|
|
Fall C, Seck A, Richard V, Ndour M, Sembene M, Laurent F, Breurec S (2012). Epidemiology of staphylococcus aureus in pigs and farmers in the largest farm in Dakar, Senegal. Foodborne Pathogens and Disease 9(10):962-965.
Crossref
|
|
|
|
|
Founou LL, Amoako DG, Founou RC, Essack SY (2018). Antibiotic Resistance in Food Animals in Africa: A Systematic Review and Meta-Analysis. Microbial Drug Resistance 24(5):648-665.
Crossref
|
|
|
|
|
Gblossi BG, Eric EA, Elise SG, Natalie G, Souleymane B, Lamine SN, Mireille D, Morgan HW (2012). Prevalence and Antimicrobial Resistance of Thermophilic Campylobacter Isolated from Chicken in Côte d'Ivoire. International Journal of Microbiology pp. 1-5.
Crossref
|
|
|
|
|
Guessennd N, Konan F, Beudje F, Tiékoura B, Ouattara D, Kouadio JN, Dosso M (2012). Sensibilité aux antibiotiques de souches d'Escherichia coli isolées des selles diarrhéiques chez des porcelets en Côte d'Ivoire. Revue Bio-Africa 10:54-61.
|
|
|
|
|
Jajere SM (2019). A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Veterinary World 12(4):504-521.
Crossref
|
|
|
|
|
Kagambèga A, Lienemann T, Aulu L, Traoré AS, Barro N, Siitonen A, Haukka K (2013). Prevalence and characterization of Salmonella enterica from the feces of cattle, poultry, swine and hedgehogs in Burkina Faso and their comparison to human Salmonella isolates. BMC Microbiology 13(1):1-9.
Crossref
|
|
|
|
|
Kadja MC, Kane Y, Viban Banah V, Kaboret Y, Bada Alambedji R (2013). Sensibilité aux antibiotiques des bactéries associées aux mammites cliniques des petits ruminants dans la région de Dakar. Annales Des Sciences Agronomiques 17(2):205-216.
|
|
|
|
|
Mama OM, Dieng M, Hanne B, Ruiz-Ripa L, Diop CGM, Torres C (2019). Genetic characterisation of staphylococci of food-producing animals in Senegal. PVL detection among MSSA. BMC Veterinary Research 15(1):1-6.
Crossref
|
|
|
|
|
Mensah SEP, Koudandé OD, Sanders P, Laurentie M, Mensah GA, Abiola FA (2014). Antimicrobial residues in foods of animal origin in Africa: public health risks. In Revue scientifique et technique (International Office of Epizootics) 33(3):987-996.
Crossref
|
|
|
|
|
Moctar M, Mouiche M, Moffo F, Tatah JF, Akoachere K, Okah-Nnane H, Mapiefou NP, Ndze V N, Wade A, Djuikwo-Teukeng F, Godelive D, Toghoua T, Zambou HR, Marc J, Feussom K, Lebreton M, Awah-Ndukum J (2019). Antimicrobial resistance from a one health perspective in Cameroon: a systematic review and meta-analysis. BMC Public Health 19:1135.
Crossref
|
|
|
|
|
Oloso NO, Fagbo S, Garbati M, Olonitola SO, Awosanya EJ, Aworh MK, Adamu H, Ayoade Odetokun I, Fasina FO (2018). Antimicrobial Resistance in Food Animals and the Environment in Nigeria: A Review. International Journal of Environmental Research and Public Health Article 15(4):1284.
Crossref
|
|
|
|
|
Ouédraogo AS, Sylvain G (2017). Emergence and spread of antibiotic resistance in West Africa: Contributing factors and threat assessment. Medicine et Santé Tropicales 2017(27):147-154.
Crossref
|
|
|
|
|
Rene KA, Adjehi D, Timothee O, Tago K, Marcelin DK, Ignace-Herve M (2014). Serotypes and antibiotic resistance of Salmonella spp. isolated from poultry carcass and raw gizzard sold in markets and catering in Abidjan, Cote d'Ivoire. International Journal of Current Microbiology and Applied Sciences 3(6):764-772.
|
|
|
|
|
Schar D, Sommanustweechai A, Laxminarayan R, Tangcharoensathien V (2018). Surveillance of antimicrobial consumption in animal production sectors of low- and middle-income countries: Optimizing use and addressing antimicrobial resistance. PLoS Medicine 15(3):1-9.
Crossref
|
|
|
|
|
Schneider K, Gugerty MK, Plotnick R, Anderson CL (2010). Poultry Market in West Africa: Overview and Comparative Analysis. Evans School Policy Analysis and Research 82:1-26.
|
|
|
|
|
Shyaka A, Kane Y, Shyaka A, Kadja MC, Kane Y, Kaboret Y, Alambedji RB (2010). Diagnostic des mammites cliniques et subcliniques en élevage bovin laitier intensif. Cas de la ferme de Wayembam (Sénégal).
|
|
|
|
|
Sidibé S, Traoré B, Sidi Y, Broulaye A, Afou D, Oumar B (2019). Antibiorésistance des souches de Salmonella gallinarum isolées en aviculture moderne en zones périurbaines au Mali. Revue d'élevage et de médecine vétérinaire des pays tropicaux 72(4):1-6.
Crossref
|
|
|
|
|
Silley P, De Jong A, Simjee S, Thomas V (2011). Harmonisation of resistance monitoring programmes in veterinary medicine: An urgent need in the EU? International Journal of Antimicrobial Agents 37(6):504-512.
Crossref
|
|
|
|
|
Somda NS, Bonkoungou OJI, Zongo C, Kagambèga A, Bassolé IHN, Traoré Y, Mahillon J, Scippo ML, Hounhouigan JD, Savadogo A (2018). Safety of ready-to-eat chicken in Burkina Faso: Microbiological quality, antibiotic resistance, and virulence genes in Escherichia coli isolated from chicken samples of Ouagadougou. Food Science and Nutrition 6(4):1077-1084.
Crossref
|
|
|
|
|
Stevens A, Kaboré Y, Perrier-Gros-Claude JD, Millemann Y, Brisabois A, Catteau M, Cavin JF, Dufour B (2006). Prevalence and antibiotic-resistance of Salmonella isolated from beef sampled from the slaughterhouse and from retailers in Dakar (Senegal). International Journal of Food Microbiology 110(2):178-186.
Crossref
|
|
|
|
|
Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, Teillant A, Laxminarayan R (2015). Global trends in antimicrobial use in food animals.
Crossref
|
|
|
|
|
Vounba P, Arsenault J, Bada-Alambédji R, Fairbrother JM (2019). Prevalence of antimicrobial resistance and potential pathogenicity, and possible spread of third generation cephalosporin resistance, in Escherichia coli isolated from healthy chicken farms in the region of Dakar, Senegal. PLoS One 14(3):e0214304.
Crossref
|
|
|
|
|
Vounba P, Yaghouba K, Ndiaye C, Arsenault J (2018). Molecular Characterization of Escherichia coli isolated from Chickens with Colibacillosis in Senegal. Foodborne Pathogens and Disease 15(8):1-9.
Crossref
|
|
|
|
|
Wall BA, Mateus A, Marshall L, Pfeiffer DU, Lubroth J, Ormel HJ, Otto P, Patriarchi A (2016). Drivers, dynamics and epidemiology of antimicrobial resistance in animal production.
View
|
|
|
|
|
World Health Organization (WHO) (2017). Integrated surveillance of antimicrobial resistance in foodborne bacteria Application of a One Health Approach.
View
|
|
|
|
|
Williams PCM, Isaacs D, Berkley JA (2018). Review Antimicrobial resistance among children in sub-Saharan Africa. The Lancet Infectious Diseases 18:e33-e44.
Crossref
|
|
|
|
|
Yao RK, Coulibaly JK, Tiekoura BK, Yapi FH, Djaman JA (2018). Molecular Characterisation of Extended-Spectrum Beta-lactamase Producing Escherichia coli Isolated from Cattle Faeces in Abidjan District, Ivory Coast. Microbiology Research Journal International 25(5):1-10.
Crossref
|
|
|
|
|
Yao RK, Julien CK, Tiécoura KB, Gueu Kpon Roméo GGB, Djaman Alico Joseph YHF (2017). Prevalence of Salmonella Strains in Cattle Breeding in the District of Abidjan (Côte d'Ivoire). International Journal of Current Microbiology and Applied Sciences 6(3):1154-1162.
Crossref
|
|