ABSTRACT
Escherichia coli O157:H7 is a well-known pathogen of man and animals and a very low infection dose is needed to propagate the infection and clinical disease. In this study, a total of 515 rectal swab samples were collected from cattle and subjected to conventional biochemical tests. Presumptive identification on Eosine Methylene Blue (EMB) yielded an overall prevalence of 83.1%. Cefixime, Tallurite, Sorbitol MacConkey Agar (CT-SMAC) test yielded 18(4.2) isolates while Indole test, Methyl Red, Voges Proskauer and Citrate utilization test (IMVIC) biochemical test showed prevalence rate of 11(61.1%) and MicrogenTM test performed on the 18 Isolates yielded a prevalence rate of 33.3%. A total of 6 of the isolates were subjected to latex agglutination test, in which 2(0.4%) were confirmed to be E. coli O157. The results of the somatic flagella antigen test performed on the 2 isolates revealed that 1(50%) belonged to E. coli O157:H7. Thus this study is therefore the first research work to confirm the presence of E. coli O157:H7 in cattle presented for slaughter in Suleja abattoir in Nigeria. Humans can contract the infection through exposure, handling and consumption of beef or animal products. Control measures are therefore necessary especially during processing and evisceration of beef, to ensure safety of cattle offal presented to the public for human consumption.
Key words: Escherichia coli O157:H7, cattle, Microgen kit, flagella staining, phenotypic identification, cattle, latex agglutination test.
E. coli are commensals that inhabit the Gastro Intestinal Tract of (GIT) of healthy animals and man. This bacterium belongs to the family Enterobacteriaceae (Aboh et al., 2015). The organism is gram negative; rod shaped with 2.0 µm long and 0.25-1.0 µm diameter can survive on a variety of substrates. It can utilize mixed acid fermentation in anaerobic condition, producing lactate, succinate, ethanol, acetate and carbondioxide (CDC, 2012). The bacterium is classified into different serotypes based on presence of pathogenic flagella antigens. Chin (2000) identified six pathotypes of E. coli namely: Diarrhoeagenic E. coli (associated with diarrhoea), Shigatoxin producing E. coli (STEC) which shared homology to the cytotoxin produced by Shigella dysenteriae, verocytotoxin producing E. coli (VTEC), Enterohemorrhagic E. coli (EHEC), associated with food poisoning, Enterotoxigenic E. coli (ETEC), Enteropathogenic E. coli (ETEC) and diffusely adherent E. coli (DAEC). The 16S rRNA based phylogenetic analysis has shown close genetic relatedness of E. coli with other members of the Enterobacteriaceae (CDC, 2012).
Human infection with shigatoxin producing E. coli O157:H7 (STECH O157) is relatively rare but the consequences could be serious, especially in the immunocompromised such as the young and the elderly (Beyi, 2017). The outcome associated with STEC O157 infection includes: Diarrhea, intense abdominal pain, hemorrhagic colitis, hemorrhagic uremic syndrome (HUS), kidney failure and eventual death (David, 2015). The infection could be transmitted directly or indirectly through fecal-oral means with organism infecting its victim through braided skin, human or animal feces, contaminated food, water or soil. Out breaks has been associated with poor hygienic measures during slaughter, evisceration and processing of beef. The detection of E. coli O157:H7 is an indicator of fecal contamination and implies presence of other dangerous pathogens which can compromise the wellbeing of consumers (Biruhtesfa et al., 2017).
E. coli O157:H7 was widely distributed in North America, along with other serotypes such as STEC O145, O26, O111 and O103, but studies have inculcated the organism to be found in processed beef in Africa and some other parts of the world (CDC, 2012). Healthy ruminants especially feedlot cattle harbor this organisms in their lower gastrointestinal tract hence, constituting major reservoir of this organisms. Cattle therefore shed these organisms in their faces, thus, disseminating this deadly disease to the environment. Other well-known vectors for the transmission of E. coli O157O:H7 includes houseflies and formites (David, 2015). Some factors are responsible for the pathogenicity of these organisms includes: Season of the year because fecal shedding rates is well known to occur during the summer and early rainfall. Also the age of cattle has shown that there is less shedding of the organisms in cattle of slaughter age than younger cattle (Beyi, 2017). The somatic antigen of E. coli O157:H7 is known to produce potent toxins which are shiga-like in nature and their distribution is based on the seasonal variability especially in healthy cattle which serves as apparent reservoirs of these organisms (Tarr et al., 2005). The organisms could be discharged in meat, offal, milk and dairy products or contaminated water apple drinks, vegetables and bovine manure (Cobbold et al., 2007).
The risk of transmission of E. coli O157:H7 to man and animals has increased overtime. The fact that low infections dose of the organism as low as ten could trigger serious infection is a signal for more researches to be conducted on this disease. This, coupled with the short incubation period of the bacterium could further exacerbate in the disease especially in the elderly and immune-compromised young individuals below five years of age (Weir and Hay, 2006).
The risk of E. coli O157:H7 from food animals has not been paid much attention in developing countries (Honise et al., 2017). There is also paucity of information regarding the epidemiology of E. coli O157:H7 in developing countries. Animals are commercially slaughtered and dressed in unhygienic conditions which compromise microbiological quality and safety of meat obtained from the animals. This consequently risks the health of the consumers. To the best of our knowledge, there are limited public health surveillance data which characterizes E. coli O157:H7 isolates from cattle presented for slaughter in Suleja abattoir, Nigeria. Our research is the first work to be conducted in the study area in order to phenotypically characterize and identify E. coli O157:H7 serotypes from rectal swabs of cattle presented for slaughter using standard bacteriological methods. This paper therefore serves as a catalyst for the need to the promotion of surveillance programs to identify sources of pathogenic E. coli from non-human origin.
Study area
Suleja is located in Suleja local government of Niger state. The city is situated at latitude 09° 31N and longitude 07° 581 E and about 20 km north of Abuja, FCT, Nigeria. It is about 100 km north east of Minna, administrative headquarters of Niger state. Suleja has a population of 216, 570 (NPC, 2006). It has a sub-humid climate, mean annual rainfall of 1640 mm and a raining season of about 7 months in the year. Agriculture which involves farming of crops and rearing of animals such as sheep and goats for sale as the need arises and also human consumption especially during festive periods.
Experimental design, sample size and sampling
A cross sectional study was conducted based on convenience observation of a selected sample of individuals from a larger population. Thereafter, each individual sample was determined by simultaneous presence or absence of disease or a causative agent of a disease and hypothesized risk factor (Dahiru et al., 2008).
The sample size was determined as described by Thrusfield (1997):
Nevertheless, 515 samples were collected in this study in order to increase the probability to capture the effect and increase the power of the study. The samples were collected from the rectum using sterilized swab sticks of the cattle presented for slaughter. The samples were placed in separate labeled sample bottles containing 8 to 9 ml Modified Tryptone Soya Broth (MTSB), kept in a cool box containing ice block were then transported to the bacteriology Laboratory of the Department of Veterinary Microbiology of the Ahmadu Bello University, Zaria, where they were processed soon on arrival. Sampling lasted for six months (that is, between July and December, 2016).
E. coli isolation and identification of O157:H7 Serotypes
All samples and media were prepared based on standard bacteriological method as indicated by the manufacturer’s instructions (Oxoid, UK). The bacterial isolation, biochemical identification and confirmation of the isolated colonies were done according to Barrow and Feltham (1993). The rectal swabs were inoculated in pre-enriched on MTSB onto Eosin Methylene Blue (EMB) agar and subsequently incubated at 24 to 48 h at 37°C. The appearance of greenish metallic sheen colonies was suggestive of E. coli. A discrete colony was then picked with sterilized wire loop, and then sub cultured onto Sorbitol MacConkey Agar suplmented with Cefixime and Tellurite (CT-SMAC Oxoid,UK) and incubated at 37°C for 24 h (Cheesebrough, 2000). The colonies on CT-SMAC were considered presumptive E. coli O157 because O157 do not ferment sorbitol. The non-sorbitol fermenters were thereafter picked and inoculated onto nutrient agar slants, incubated at 37°C for 24 h and stored in the refrigerator for further biochemical tests.
Stocked isolates suspected to be E. coli were further verified using conventional biochemical tests as described by Harrigan (1988) on the basis of indole production and motility with SIM medium (Merch, Germany), citrate utilization with simmons citrate agar (Merck, Germany), methyl red and vogues-proskauer using MR-VP medium (Merk, Germany) and urease production using urea agar (Oxoid, UK).
MicrogenTM confirmation of E. coli isolates
Commercially available biochemical test strip (MicrogenTM ID, UK) was used to further confirm isolates suspected to be E. coli based on the results of the conventional biochemical tests (OIE, 2008). The isolates to be inoculated were grown on selective media (CT-SMAC Oxoid, UK) using a wire loop, 3 to 4 discrete colonies were emulsified in normal saline solution (0.85%) and adjusted to McFarland turbidity standard of 0.5. The wells of the individual substrates sets were exposed by cutting the end tag of the sealing strip and slowly peeling it back. Using a micropipette with sterile micropipette tips, 100 µl of the bacterial suspension was added to each well in the set. Mineral oil was then used to overlay the substrates in wells 1, 2 and 3 using a Pasteur pipette. The inoculated rows were then re-sealed and properly labeled at the end of the tag with the specimen identification number followed by incubation at 37°C for 24 h. On 24 h incubation period, the sealing tape on the test strips peeled back and evaluated. Results were then recorded on a report form as positive or negative by comparing them with a colour reference to the table of reactions provided. In addition to well 8 and (Indole production), two drops of Kovac’s reagent were added and the results evaluated within 2 min of adding the reagent. To well 10 (Vogues – proskauer reaction), 1 drop each VP1 and Vp2 reagents were added and the results evaluated within 15 to 30 min, after addition. To well 12 (Tryptophan Deaminase), 1 drop of TDA reagent was added and the results were evaluated immediately.
In the interpretation of the results, the octal coding system was adopted with each group of the three reactions producing a single digit of the code. Using the results recorded on the report forms, the indices of the positive reactions are circled and the sum of these indices in each group of these three reactions formed a code of four numbers. This code obtained was then entered into the computer-aided identification package and the resulting organisms and its percentage probability was recorded.
Serotyping of E. coli O157:H7 (latex agglutination test)
The isolates that were confirmed as E. coli using the MicrogenTM were further serotyped using bacterial agglutination test using a commercial latex kit for E. coli 0157:H7 (Wellcolex E. coli O157:H7 Kit, Oxoid, UK) which is a rapid Latex Agglutination test for confirming non-sorbitol fermenting colonies and H7 somatic flagella antigen (OIE, 2008).
Detection of E. coli O157
Fresh cultures grown at 37°C for 24 h on sorbitol MacConkey agar (Oxoid, UK), were used for this test. 40 µl of saline was placed in two circles on a reaction card, using a micropipette. With the help of a mixing stick provided, sufficient growth just enough to cover the blunt end of the stick was removed from the plate and emulsified in the saline by rubbing with the flat end of the stick, mixing it thoroughly without damaging the surface of the card. This was done for the second circle on the reaction card already containing saline after which mixing sticks were immediately discarded. For each of the test sample, one drop of O157 test latex was placed in one circle (with the emulsified culture) and one drop of O157 control latex in the other circle by holding the dropper bottles vertically in order to dispense on accurate drop. A mixing stick was then used to mix the contents of the circles, spreading the latex carefully over the entire area of the circle. The card was then rocked slowly for 30 s and observed for agglutination. The used reaction card was then discarded for safe disposal. The agglutination of O157 test latex accompanied by a lack of agglutination of the control latex on observation was used to indicate the presence of O157 antigenic the culture under test. Absence of agglutination both O157 test control reagents was used to indicate the absence of O157 antigen in the test culture (OIE, 2008).
Detection of H7 (Flagella Staining)
Test cultures that were positive for O157 antigen were further tested for H7 antigen by sub culturing in Modified Tryptone Soya Broth (MTSB) Oxiod, UK at 37°C for 24 h. For each sample, 40 ul each of MTSB-grown cultures were placed in two circles on a reaction card, followed by placing a drop of the H7 test latex in one circle (with the broth culture) and a drop of H7control latex in the other circle. The contents of the circles were then thoroughly mixed using a mixing stick, carefully spreading the latex over the entire areas of the circle. The card was then slowly rocked for 30 s and observed for agglutination after which the card was discarded. Agglutination of the H7 test latex accompanied by a lack of agglutination of the control latex on observation was used to indicate the presence of H7 antigen while the absence of agglutination in both reagents was indicative of absence of H7 in the test culture (OIE, 2008).
Data analysis
The results were presented using simple descriptive statistics involving percentages, tables and charts (CDC, 2012).
Various reactions to confirm E. coli O157 and E. coli O157:H7 are shown on Figures 1 to 3. Of the 515 rectal swab samples analysed, 428 (83.1%) were presumably positive for E. coli, with characteristic greenish metallic sheen. Sub culturing on cefixime, tellurite sobitol MacConkey (CT-SMAC) agar revealed 18 (4.2%) as non-sorbitol fermenting strains of E. coli O157:H7. However, 87 (20.3%) of the 515 samples did not yield any significant bacterial growth (Table 1). Table 2 showed the results of the biochemical tests (IMVIC), Microgen TM, latext agglutination test and flagellar staining of the 18 isolates tested for IMVIC, 11(61.1%) were identified as E. coli. The MicrogenTM GMA + B-ID system, UK showed that from the 18 isolates tested, 6(33.33%) were positive for MicrogenTMtest. The somatic and flagella staining for 6 isolates yielded 2 (0.4%) of E. coli 0157 while from the 2 of the isolates tested for flagella staining, 1 (50%) yielded positive results.
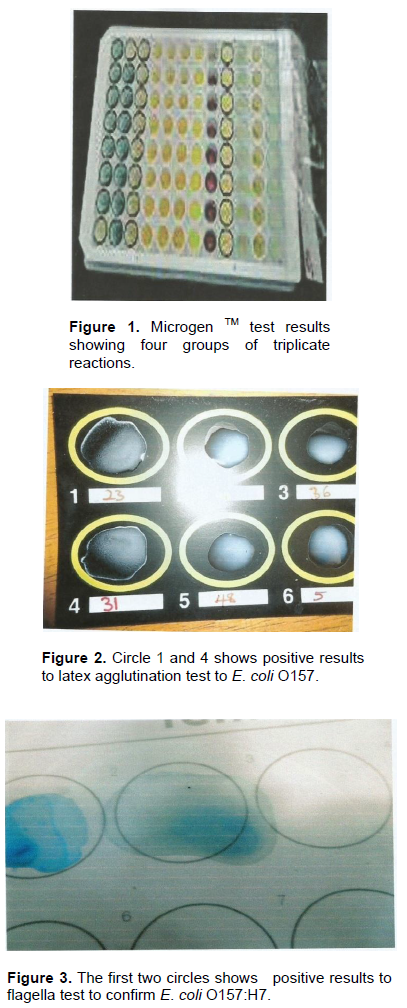
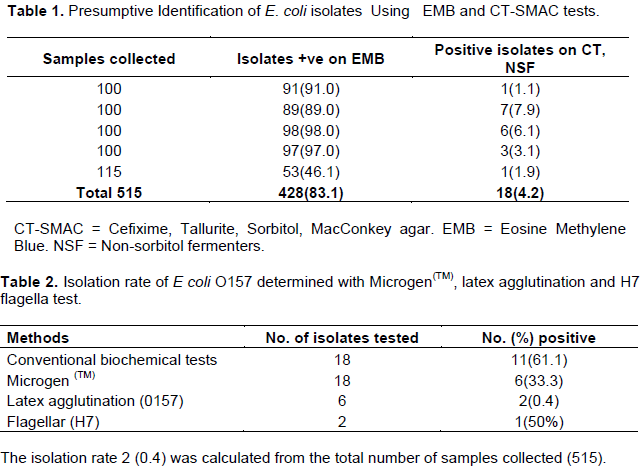
Figure 1 shows the Microgen TM test of the groups of triplicate reactions of E. coli isolates. Figure 2 shows that the 4th circle which contains the positive results to Latex Agglutination test confirming E. coli O157. Figure 3, on the other hand shows the positive results to flagella staining, with the first two circles dissipating the positive results of presence of E. coli O157:H7.
The presumptive prevalence of 428(83.1%) E. coli isolates signifies a high isolation rate of E. coli as indicated in our studies. The high prevalence rate could be associated with poor hygienic practices right from the abattoir, during handing or transportation of the carcasses to the market. E. coli is a normal commensal of gastrointestinal tract of cattle and man. This observation has been also been made in other animals species including sheep, goats, pigs, birds and non-human primates (Nuno et al., 2011; Rosa et al., 2017). The organism which was thought to be non-pathogenic for several years however has become highly adapted to cause diarrhoea, septicaemia, meningitis, urinary tract infection, wound infections in several animal species and humans. E. coli is well known to cause infection not only in immuno compromised patients but also in situations of impaired gastrointestinal barriers (Keskimaki et al., 1997). The isolation of E. coli from cattle in Suleja abattoir is therefore of public health significance.
The isolation rate of E. coli O157:H7 obtained from this study was found to be 0.4%. This may seem low but could present a more challenging effect especially in cases involving human exposure, since a very low dose of the pathogen is required to establish infection (Podolak et al., 2010). The low isolation rate of E. coli O157:H7 at 0.4% observed in our present study concurs to previous reports of Tutenel et al. (2002) with slight variation, who reported low isolation rate of 0.7 and 1.2% from rectal swabs of slaughtered cattle in Poland and Finland respectively. The low prevalence of E. coli O157:H7 in cattle suggests low infection rate in the cattle population studied. This finding can be extrapolated for a larger cattle population in Nigeria with caution. Although it is difficult to trace back the herds’ origin of the cattle presented for slaughter, it is reasonable to assume that the abattoir in Suleja presents a wide catchment area where animals are brought for slaughter across the country. This finding can be confounded by the fact that cattle could be brought from big or small herds; or even infected or uninfected herds. These calls for large scale studies to determine the prevalence and distribution of E. coli O157:H7 under different cattle production systems in Nigeria.
However, Elder et al. (2000) reported a higher isolation rate of 27.8% from rectal swabs of slaughtered cattle in USA; and Dahiru et al. (2008) reported a very high isolation rate of 53% from fresh beef in Kano state, Nigeria. On the other vein, the isolation rate of 2.8% for E. coli 0157: H7 was reported from human feces in Ile-Ife, Nigeria (Odetoyi et al., 2016). These reports have lent credence to the high and increasing endemicity of E. coli. O157:H7 in different geographical locations including abattoirs and slaughter houses globally. However, the variations in the isolation rates may be attributed coupled with variations in sampling methods and isolation techniques used, as well as the distribution and seasonal variation of the organisms from one geographical location to another or within different countries.
The recto anal junction of cattle serves as the principal predilection site for predilection of E. coli O157:H7 (Rosa et al., 2017). Cattle have been known to be established as natural reservoir for the dissemination of E. coli O157:H7. Cattle play significant role in the epidemiology of human and animal infection, In the present study, the presence of E. coli O157:H7 in cattle presented for slaughter may suggest high possibility of unwholesome practices, leading to the contamination of carcasses by fecal materials during slaughter and processing of beef, which suggests that the current available processing procedures at the abattoir are not reliable to prevent fecal contamination during slaughter. It is therefore necessary that government should intensify regulatory efforts to curb this menace. Epedemiological studies have shown that cattle are healthy carriers of E. coli O157:H7 and that these organisms produce potent toxins which could be shed in feces of apparently healthy cattle (OIE, 2008). Therefore, the findings indicated in our studies are therefore very significant. Prevention and total eradication of this disease is important in improving cattle manage-ment practices and safeguarding the public.
Though, the study revealed low isolation rate of E. coli 0157:H7 at 0.4%, it also exposes virulent strains of these isolates. This therefore sends a warning signal to relevant regulatory authorities in Nigeria that the disease is very much around. E. coli O157:H7 may spread from cattle to humans. It therefore calls for rampant public health awareness campaign and development of relevant disease control strategies in Nigeria.
The authors wish to declare that they do not have any conflict of interests.
REFERENCES
|
Aboh EA, Giwa GJ, Giwa A(2015). Microbial Assessment of well water in Samaru, Kaduna State. Ann. Afr. Med. 14(1):32-38.
Crossref
|
|
|
|
Barrow GI, Felthan RKA (1993). Cowan and Steel's manual for the Identification of Medical Bacteria 3rd ed.), Cambridge University Press. 128-146:222-238.
Crossref
|
|
|
|
|
Beyi AF (2017). Prevalence and antimicrobial Susceptibility of E.coli O157: H7in beef cattle at butcher shops and restaurants in Central Ethiopia. BMC Microbiol. 217:17-49.
|
|
|
|
|
Biruhtesfa A, Degmawi P, Mesele A, Genene T, Dereja H, Sufafel K, Kebede A (2017). Occurrence of E.coli O157: H7 in cattle feces and contaminated carcasses in various contact surfaces in Abbattoir and Butcher shops in Nawasse, Ethiopia. BMC Microbiol. 17:34-35.
|
|
|
|
|
Centres for Disease Control and Prevention (CDC) (2012). Human illness caused by E. coli 0157: H7 from food and non-food sources. CDC 27/7 saving lives and protecting people. pp. 3-4.
|
|
|
|
|
Cheesebrough M (2000). District Laboratory Practice in Tropical Countries. Part 2, Cambridge University Press. pp. 381-407.
|
|
|
|
|
Chin J (2000). Control of Communicable diseases manual, 17th Ed. An official report of the American Public Health Association. Washington D.C. American Pub. Health Assoc. pp. 90-91.
|
|
|
|
|
Cobbold RN, Hancock DD, Rice, DH, Berg J, Stillborn R, Houdec CJ, Besser JE (2007). Recto-anal Junction colonisation of feed lot cattle by Escherichia coli0157: H7 and its association with super shedders and excretion dynamics. Appl. Environ. Microbiol. 73:1563-1568.
Crossref
|
|
|
|
|
Dahiru M, Uriah N, Enabulele SA, Shamsudeen U (2008). Prevalence of E. coli O157: H7 in fresh and roasted beef in Kano city, Nigeria, Bayero J. Pure Appl. Sci. 1(1):39-42.
|
|
|
|
|
David RS (2015). Prevalence of E. coli O157: H7 in beef cattle and slaughter beef carcasses in retail shops in Ethiopia. ASM, Washington DC. Micro. Bioshere. EHEC-0006-2013.
|
|
|
|
|
Elder RO, Keen JE, Siragusa GR, Barkocy-Gallagher, GA, Kooh M, Maraier U, lecgried WW (2000). Correlation of Enterohaemorrhagic E. coli 0157: H7 prevalence in faeces, hides and carcasses of beef cattle during processing: Proceedings of the beef cattle during processing. Proc. Natl. Acad. Sci. 97:2999-3003.
|
|
|
|
|
Honise L, Punja N, Num S, Nelson D, Hiislop N, Gosselin G, Stashko N, Ditrich C (2017). E.coli O157: H7 infections associated with contaminated pork products. Can. Commun. Dis. Rep. 43(1):21-24.
|
|
|
|
|
Keskimaki M, Ikäheimo R, Kärkkäinen P, Scheutz F, Puohiniemi RL, Siitonen A (1997). Shiga toxin producing Escherichia coli serotype OX3:H21 causing hemolytic uremic syndrome. Clin. Infect. Dis. 24:1278-1279.
Crossref
|
|
|
|
|
NPC-National Population Commission (2006). Archived data on 2006 Nigerian Population Census. pp. 55-56.
|
|
|
|
|
Nuno S, Igrias G, Goncalves A, Poeta P (2011). Commensally gut bacteria: distribution of Enterococcus Species and prevalence of Escherichia coli phylogenetic groups in animals and humans in Portugal. Ann. Microbiol. 62(2):449-459.
|
|
|
|
|
Odetoyi BW, Hofmann J, Aboderin A, Okeke IN, (2016). Diarrheoagenic E.coli in mother and child pairs in Ile-Ife, south western Nigeria. BMC Infect. Dis. Ser. 16:28-30.
|
|
|
|
|
OIE-Office of International Epizootics (2008). Vero cytotoxigenic Escherichia coli. Terres. Man. Chapter 2.9.11. pp. 1295-1298.
|
|
|
|
|
Podolak I, Galanty, A, Sobolewska D (2010). Saponins as cytoxic agents: A review. Phytochem. Rev. 9(3):425-474.
Crossref
|
|
|
|
|
Rosa A, Woynshet H, Akefete T, Ashenafi F, Getalum EA, Badeso m, Fanos T, Geloye K, Takele B, Teriku J, Bayene LDC, Eric C Bruno MG (2017). Prevalence of E. coli in beef cattle slaughtered in Ethiopia. BMC Infect. Dis. 17:21.
|
|
|
|
|
Tarr PT, Gordon CA, Chandler WL (2005). Shiga-toxin producing E. coli and haemolytic uremic syndrome. Lancet 365:1073-1086.
|
|
|
|
|
Thrusfield M (1997). Veterinary Epidemiology. 2nd Edition, Blackwell Scientific Publications, Oxford. pp. 53-55.
|
|
|
|
|
Tutenel AV, Perard D, Uradzinski J, Jo-wiki E, Pastuszczak M, Van Hende J, Uyttendaele M. Debevere J. Cheasty T, Van Hoof J, De-zutter (2002). Isolation and characterization of enterohaemorrhagic Escherichia coli 0157:H7 from cattle in Belgium and Poland. Epidemiol. Infect. 129(1):41-47.
Crossref
|
|
|
|
|
Weir E, Hay K (2006). E. coli—sporadic case or an outbreak?. Can. Med. Assoc. J. 174(12):1711.
Crossref
|
|


