ABSTRACT
Analysis of water bodies, such as rivers and lakes can provide an insight into their intrinsic composition and potential impact on the environment. Water samples collected from four designated sites in Idah River, were assessed for their physicochemical parameters and microbial diversity using standard procedures. The temperature from all sites was 26.00°C and the pH ranged from 6.93 to 7.08. Turbidity values ranged between 32.00 and 38.00 NTU, while dissolved oxygen ranged between 6.28 and 9.28 ppm. Heavy metals, such as Selenium and Arsenic (with peak values of 0.10 and 0.09 ppm, respectively) were detected in the river. However, dissolved oxygen, arsenic and turbidity values across all sites exceeded the maximum limit set by World Health Organisation and the Standard Organisation of Nigeria. The total heterotrophic bacterial counts showed excessive bacterial load from all sample sites, while pathogenic bacteria, including Escherichia coli, Klebsiella and Shigella species, were isolated from regions with intense anthropogenic activities along the river, indicative of microbial pollution. Fungal studies identified the presence of Fusarium, Aspergillus and Trichoderma species as the most abundant in the river. Obtained results showed that Idah River is exposed to heavy metal seepage and subject to microbial contamination. Therefore, continuous monitoring and better management of the river body is recommended to prevent disease outbreak.
Key words: Aquatic ecosystems, Idah River, microbial diversity, physicochemical analysis.
Water can be sourced from water bodies such as rivers, boreholes, lakes, springs and other large water bodies. However, the quality of water bodies can be adversely affected by man-made activities. For example, pollution of freshwater bodies such as rivers (e.g. Idah River, Kogi State), streams, lakes and ponds are mostly experienced due to industrial discharge, municipal waste disposal and surface run-off (Akaniwor et al., 2007). Anthropogenic activities, such as discharges of domestic waste, untreated waste from sewage treatment plants, plastic materials, disposal of personal care products and household chemicals, improper disposal of car batteries, construction activities, mining activities, and pilgrim activities constantly deteriorate the water quality of rivers (Environmental Pollution Centers, 2018). Such deterioration in water bodies include an alteration in pH, increased turbidity, higher content of total dissolved solids and metals, as well as a higher risk of such water body hosting water-borne pathogens (Shaji et al., 2009; Ananthakrishnan et al., 2012). Water-borne pathogens pose a great health risk to humans, animals and plants, most especially, infants, young children under the age of ?ve and immunocompromised individuals (EPA, 2016; WHO, 2014).
River Niger, at Idah, is an extension of the two major Nigerian rivers; River Niger- after which the country Nigeria is named, and River Benue, after their confluence in Lokoja. The Idah River is located between latitude 7°6'1"N and longitude 6°42'23"E (Figure 1) and it serves as a boundary between Kogi and Edo states. The temperature of the water ranges between 22 and 31°C (which greatly depends on the season). It is relatively turbid and has a slightly alkaline pH between 7.5 and 7.8 (depending on the sampling location). The highest water levels are usually recorded between August and September, while the lowest water level is usually recorded between March and April (Adeyemi, 2010).
Although, microbial communities represent a fundamental part of aquatic ecosystems, and are of great importance for matter and energy flux (Kavka et al., 1996), little is known about the microbial biodiversity of River Niger (Idah Axis), despite its popularity in Nigeria and beyond. Local communities at the different axis of River Niger bank exploit the water for the fishery, aquatic medicinal plants, and domestic purposes, such as cooking, drinking, bathing, washing utensils and clothes, while the water is exposed to both human and animal waste, discharge of untreated industrial and domestic wastewater, runoff and dead organic plants and animals. Studying the unique diversity and functions of microbes in their ecological niche, as well as establishing the factors that affect them would aid in unraveling the role they play in animal, human, plant health, and in their environment (Marchesi, 2017).
This study is therefore aimed at evaluating the total microbial content of water at different points of River Niger at Idah axis, using a conventional approach. In addition, the study seeks to link the community present to the physiochemical factors that characterize the river, in a bid to proffer necessary recommendations to avert infectious disease outbreaks.
Study site
Idah is a town in Kogi State, in the middle belt region of Nigeria and situated on the east bank of Niger River. It has a land mass of 36 km2 and located on latitude 7°06'48.42"N and longitude 6°44'19.18" E. Its human population was around 79,815 as at 2006 (National Population Commission, 2006). It has a tropical savannah climate with a wet season between late March and early November, and dry season between early November and late March. Idah River is exposed to water pollutants, such as chemical waste from farming activities, as well as discharge from gullies and streams, including the Inachalo River. The locals consume the water for domestic and dry farming activities. During rainy season, run-off washes domestic and municipal sewage, abbatoir effluents and remnants of open defecation of both human and animals into the river.
Sample collection
Towards the end of the dry season in February 2020, a total of 20 water samples were aseptically collected in replicates from 4 selected sites in the river (designated Docking Point A, Docking Point B and the Idah Axis - Midstream and Confluence Area); all characterized by different levels of anthropogenic activities (Figure 1). Samples for bacteriological, mycological and physicochemical analyses were collected, at a depth of 20 cm below the water surface and against the water current into 250 mL sterile clean bottles (Ademorati, 1996). The temperature and water pH were monitored in situ using a mercury glass thermometer and a pH meter (ROHS Model), respectively before sampling (APHA, 2005). Collected samples were preserved in an icebox and transported immediately to the laboratory for microbiological and physicochemical analyses.
Physicochemical examination of water samples
Water samples placed on icepacks were transported to Soil Microbiology Laboratory at the International Institute of Tropical Agriculture, Ibadan, Oyo State, Nigeria within 24 to 48 h, for physicochemical analysis. In addition to pH and temperature (recorded in situ), a total of eighteen physicochemical parameters were evaluated. These included dissolved oxygen, specific ions (calcium, magnesium, potassium, sodium, iron, copper, nickel, arsenic, and selenium), turbidity, nitrates, sulphates, total dissolved solids, total nitrogen and electrical conductivity using titrimetric, colorimetric or spectrophotometric assays (Feng et al., 2009; Ologbosere et al., 2016; Adedire et al., 2021).
Isolation and identification of bacteria
Using aseptic techniques, four-fold serial dilution of each water sample was made, and isolation of bacteria from a serially diluted sample (1 mL) was done in Petri dish through the pour plate method on Nutrient Agar, prepared according to manufacturer’s specification. The plates were incubated at 37°C for 24 h. The mean total viable count of the isolated bacterial colonies was enumerated and recorded. Colonies with distinct morphological differences were randomly picked from the nutrient agar plates and repeatedly streaked on fresh, sterile agar plates (Nutrient Agar (Lifesave Biotech, USA), MacConkey Agar (Lifesave Biotech, USA), Eosin Methylene Blue Agar (Lifesave Biotech, USA), Salmonella-Shigella Agar (Lifesave Biotech, USA), and Centrimide Agar (Oxoid, UK) and subsequently incubated at 37°C for 24 h. Pure cultures were stored in the refrigerator at 4°C for further characterization and analysis. Pure cultures of isolates were characterized and identified using their macroscopic and microscopic characteristics, as well as biochemical tests (Catalase, Oxidase, Centrimide, Citrate Utilization, Methyl Red, Voges Proskauer, Urease, Gelatin Hydrolysis, Motility, Sugar Fermentation: Glucose, Lactose, Maltose, Sorbitol) as described by Cheesebrough (2000) and following the Bergey’s Manual of Determinative Bacteriology (Genhardt et al., 1994).
Isolation and identification of fungi
Fungi were isolated through serial dilution (10-2 and 10-4) method using sterile distilled water. One milliliter of each dilution was aseptically transferred into Potato Dextrose Agar (Lifesave Biotech, USA) media plates supplemented with Streptomycin (0.03 g/L) to inhibit bacterial growth (Hageskal et al., 2006). The plates were incubated at room temperature (25 ± 3.00°C) for 5 to 7 days. Using a flamed inoculating needle, the edge of each growing colony was picked and slides of the different colonies were made. A drop of Lactophenol cotton blue stain was added to the prepared slides, covered with a coverslip before microscopic examination using 10x and 40x magnifications to observe the microscopic features (hyphal characteristics, shape of sporangia, conidia and spores) of each isolate. Fungal colonies were also identified using cultural (macroscopic) characteristics (colony texture, elevation, chromogenesis/pigmentation, opacity and size) (Watanabe, 2002).
Statistical analysis
Data from physicochemical determinations and bacteriological plate counts were analyzed using Duncan’s Multiple Range Test using SPSS (version 25.0). Mean occurrences of bacterial and fungal isolates were tested using Tukey Pairwise comparison of grouping at a 5% level of probability using SPSS (version 25.0).
Physicochemical characteristics of water samples from different sites of Idah River
The results of the physicochemical analyses of water samples collected from four different sites in Idah River are shown in Table 1. The observed water quality of the river was compared to acceptable standards reported by the World Health Organization (WHO, 2011) and Standard Organization of Nigeria (SON, 2007). The pH of water samples taken from different sampling points within Idah River was significantly different. However, the pH of Docking point A (DPA) and Idah Axis Confluence (IAC) water samples, as well as Docking point B (DPB) and Idah Axis Midstream (IAM) water samples, respectively were not significantly different. The pH ranges obtained also showed that IAM had the highest mean pH value of 7.08, which was quite similar to that obtained at Docking Point B (7.07). The lowest pH recorded was at the IAC site (6.93) (p < 0.05).

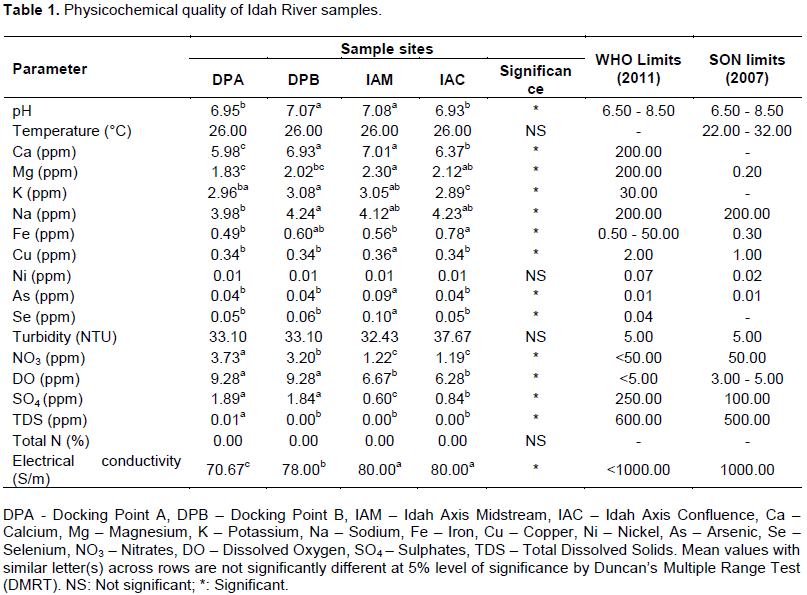
The recorded temperature was uniform across all sampled sites at 26.00°C. The mean values of calcium (7.01 ppm), magnesium (2.30 ppm), copper (0.36 ppm), arsenic (0.09 ppm) and selenium (0.10 ppm) were all found to be the highest in IAM. DPA had the lowest mean values for calcium (5.98 ppm), magnesium (1.83 ppm), sodium (3.98 ppm) and iron (0.49 ppm). The mean values of nickel, at 0.01 ppm, were uniform across all sampled sites. The highest potassium (3.08 ppm) and sodium (4.24 ppm) were recorded at DPB, and they were significantly higher than values recorded from other locations. Copper had a uniform value (0.34 ppm) at three sites: DPA, DPB and IAC areas. Turbidity mean values ranged from 33.10 NTU (at DPA and DPB) to 37.67 NTU (at IAC) and the values were not significantly different from one another. Nitrates, dissolved oxygen and sulphates were all found to be the highest in DPA with mean values of 3.73, 9.28 and 1.89 ppm, respectively.
Bacteria distribution and frequency
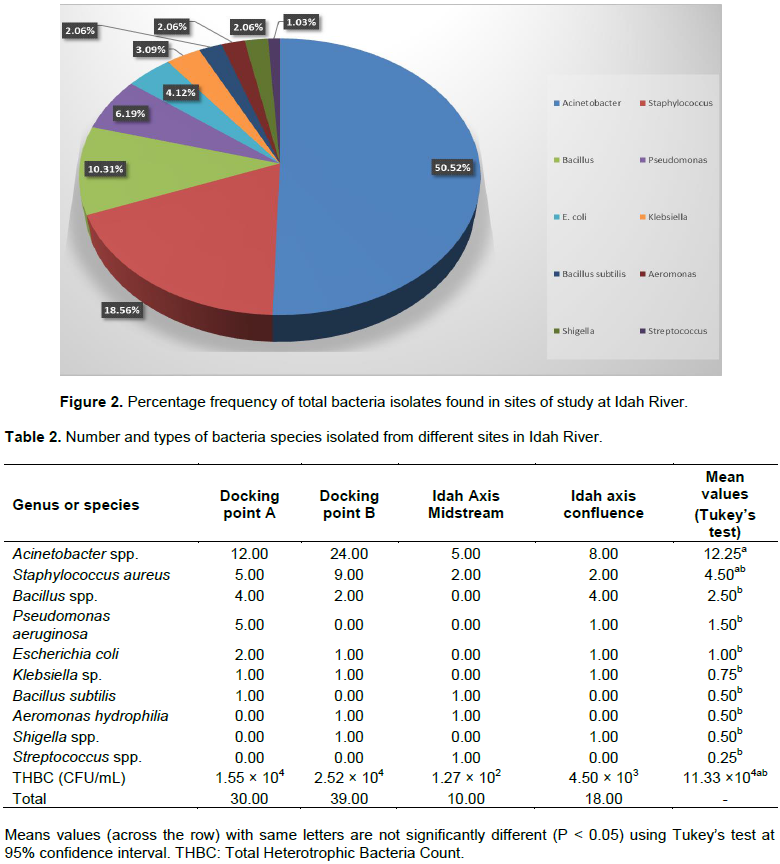
The total heterotrophic bacteria counts (THBC) of bacteria are seen in the four sites shown in Table 1. The highest microbial load was recorded at both DPA and DPB with mean values of 1.55 × 104 and 2.52 × 104 CFU/mL, respectively. The lowest microbial load was recorded at IAM with a mean population of 1.27 × 102 CFU/mL. Ten genera of bacteria were identified from a total of 97 isolates. These genera included Acinetobacter species, Staphylococcus species, Bacillus species, Pseudomonas species, Escherichia coli, Klebsiella species, Bacillus subtilis, Aeromonas hydrophilia, Shigella species and Streptococci species. Out of these ten species, Acinetobacter spp. had the highest percentage frequency of occurrence (50.52%), followed by Staphylococcus (18.56%) and Bacillus spp. (10.31%) (Figure 2). Across all sampled sites, Acinetobacter, Staphylococcus and Bacillus spp. were the most predominant. The least dominant microbes from each sample site were B. subtilis (in DPA and IAM), A. hydrophilia (in DPB and IAM), Shigella spp. (DPB and IAC) and lastly, Streptococcus spp. (in IAM only). Cumulatively, Acinetobacter spp. had a mean microbial prevalence of 12.25 and was the most predominant bacteria (P<0.05), while Streptococcus spp. was the least with a cumulative mean value of 0.25. Also, the two docking points A and B had the highest number of bacterial isolates with 30.00 and 39.00, respectively, while the least number of bacterial isolate (10) was observed at the Idah midstream area (Table 2). Concerning the total viable THBC observed in water samples collected from the four sample sites, bacteria colonies ranged from 1.27 × 102 CFU/mL (IAM) to 2.52 × 104 CFU/mL (DPB).
Fungi distribution and frequency
Seven fungal genera were identified from a total of 23 fungal isolates. These included Aspergillus flavus, Aspergillus niger, Penicillium species, Fusarium species, Cladosporium species, Trichoderma species and Curvularia species. Table 3 shows their distinct morphological differences and microscopic characteristics with 2 of the fungi species belonging to Aspergillus genus. Fusarium spp. had the highest percentage occurrence (39.13%), followed by Trichoderma spp., A. flavus and Penicillium spp. (all at 13.04%) (Figure 3). Fusarium spp. had the highest microbial incidence across all sites with a mean value of 2.25 (P<0.05), and the least occurring isolate being Curvularia spp. with a cumulative mean of 0.25. Also, DPA had the highest number of fungi isolates (9.00), followed by DPB (7.00) while the IAM area had the lowest number of fungi isolates 2 (Table 4).
From the analysis of the physicochemical features of water samples, recorded pH values were within a narrow range of 6.93 to 7.08. This fell within the acceptable range according to the standards set by WHO (2011) and SON (2007), thus making it suitable for aquatic life. This observation was in agreement with the findings of Adesakin et al. (2020) who reported a similar pH range for domestic water sources in Zaria, Nigeria. The pH values observed in this study could be attributed to the major soil type in the area or the buildup of organic materials from runoff, as described by Taiwo et al. (2020).
The pH in water bodies is among the major physiological factors that play a critical role in shaping microbial structures and other biological activities in water (Adesakin et al., 2020). There appears to be a correlation between the microbial population observed across all sample sites and the pH of Idah River. All isolated microbes were neutrophilic in nature and this may be related to the near-neutral pH of the river body, encouraging the proliferation and prevalence of neutrophils.
The temperature of any water body determines the proliferation and survival rate of microorganisms (Bouzid, 2016). In this study, the temperature value of 26.00°C in water samples could be regarded as optimal for the growth of heterotrophic mesophilic bacteria and fungi. However, this was within the standard permissible temperature limit of SON (2007) set for aquatic water life. Igbinosa et al. (2012) also observed a similar temperature range of 26 to 27.2°C in Shanomi creek, in the Niger Delta region. In addition, Nwoko et al. (2015) in their report on the assessment of seasonal physicochemical parameters of Oguta Lake in Nigeria, observed a similar temperature range.
The calcium (Ca) levels in water samples collected from different points along the river, was significantly different from each other and ranged from 5.98 ppm (DPA) to 7.01 ppm (IAM); however, these levels were below the permissible limit (200.00 mg/L) recommended by WHO (2011). As such, there might be no detrimental effect on aquatic life and on humans who ingest foods harvested from the river. Magnesium is generally associated with calcium in water bodies and with a concentration usually lower than that of calcium (Venkatasubramani and Meenambal, 2007). Magnesium values observed in this study support this statement as they ranged from 1.83 ppm (DPA) to 2.30 ppm (IAM).
The potassium and sodium values ranged from 2.89 ppm (IAC) to 3.08 ppm (DPB) and 3.96 ppm (DPA) to 4.23 ppm (IAC), respectively. Many water bodies have sodium concentrations well below 50.00 mg/L (Ikhuoriah et al., 2016). Copper levels ranged narrowly from 0.34 to 0.36 ppm and they were notably lower than what Kuz’mina and Ushakova (2007) reported in their metal assessment of fishponds (10.6 ppm). The heavy metal, Nickel is known to be associated with gastrointestinal irritation without inherent toxicity (Hammer and Hammer, 2003). In this study, Nickel value was recorded as 0.01 ppm across the four different samples, which was within the acceptable limit in WHO (2011) recommendations, hence, the river appeared devoid of the heavy metal. These values were contrary to the much higher values reported by Okereke (2014), who assessed the physicochemical properties of the Ihuku River (0.07 - 0.082 mg/L).
Arsenic is a metalloid widely distributed in the earth and usually found in natural groundwater (Thi et al., 2009) but its concentrations vary based on the geological formations, weathering processes of rocks, microbial activities, leaching or other anthropogenic activities such as mining and application of pesticides (Katsoyiannis et al., 2004; Oremland and Stolz, 2003). The metal has no known biological function and is extremely toxic in high concentrations. The arsenic mean values in this study were slightly above the WHO permissible limit, with the highest being 0.09 ppm in the IAM area. This can pose certain risks to consumers of water from the river, as this metal has been characterized as a carcinogen. Regular consumers of contaminated water and contaminated aquatic foods may be at risk of arsenic buildup in the body. Selenium is an essential micronutrient found in groundwater as well but toxic at elevated levels. Its concentration levels usually rise when there is an occurrence of industrial emissions or mining activities (Fernandez-Martinez and Charlet, 2009). Selenium recorded for the different water samples were relatively close to recommended WHO limit (0.05 mg/L) except for the IAM sample, which showed a high value of 0.10 ppm.
Turbidity is the degree of clarity or cloudiness of water and it is evaluated by the presence of suspended solids in the water (Health Canada, 2012). The mean turbidity values obtained from all water samples ranged from 32.43 to 37.67 NTU. This raises another concern, as the values obtained exceeded recommended limits by WHO and SON, thereby rendering it unfit for drinking. Suspended solids in water promote the growth of microorganisms and as such, high turbidity is often correlated with a high presence of disease-causing microorganisms (Shittu et al., 2008). The high turbidity values obtained are clear indicators of Idah River pollution. The concentrations of the nutrients (nitrates and sulphates) were within the acceptable limits for both WHO and SON standards. Therefore, they did not pose a serious water quality issue.
Dissolved oxygen (DO) is a measure of oxygen levels dissolved in an aqueous solution, which plays a major role in the biological activities of cultured organisms (Murphy, 2005). It is an important criterion for assessing quality, as it provides details on pollution levels, metabolic activities of microorganisms and nutrient availability (Premlata, 2009). The DO concentration range for the four sites: DPA (9.28 ppm), DPB (9.28 ppm), IAM (6.67 ppm) and IAC (6.27 ppm) were all higher than the statutory permissible limit (5 mg/L) necessary for aquatic life. Although, slightly above the permitted threshold, there have been higher concentrations of DO reportedly used for agricultural purposes in brackish aquaculture (Ayedun et al., 2011). Also, microorganisms use DO for the decomposition of organic materials at the bottom of water bodies. Low DO may lead to an anaerobic environment, resulting in a bad odour of water (Adekunle et al., 2007).
Electrical conductivity is defined as a measure of the degree of ions in an aqueous solution or the capacity of water to pass electrical flow. The source of these conductive ions could be from dissolved salts and inorganic materials such as chlorides, carbonate compounds or sulfides (Miller et al., 1988). The range of electrical conductivity values recorded from all sample sites was within the acceptable limit reported by WHO (2011). This may imply that the river receives low amounts of dissolved salts and inorganic substances (Kidu et al., 2015).
Results also showed that lesser concentrations of calcium, magnesium, potassium, nitrates and sulphates were recorded in IAM and IAC sample sites. This might be connected to the continuous movement of water, thereby diluting the concentrations of organic and inorganic wastes that might have been introduced at the docking sites of the river.
The total heterotrophic bacteria counts of bacteria exceeded the WHO’s stipulated standards for water bodies. Notably, the high population of bacteria seen at the two docking points could be due to the fishing and other observed anthropogenic activities occurring around the Idah riverbank. The high abundance of bacteria isolated from these two areas could also be linked to the bathing activities, washing and sewage run-offs, which was noticed at the riverbank, and is most likely to transmit an array of infectious diseases (Anyanwu and Okoli, 2012).
In the IAM area, the highest arsenic and selenium values were recorded, which were above the WHO limit. This might be indicative of toxicity, and could be the reason for the low heterotrophic bacterial plate count and the low number of bacteria and fungi isolated (Leon et al., 2018).
The bacteriological analysis showed that Gram-negative bacteria were predominant amongst the bacterial isolates from Idah River. The microorganisms isolated in order of prevalence were Acinetobacter spp., Staphylococcus aureus, Bacillus spp., Pseudomonas aeruginosa, E. coli, Klebsiella spp., B. subtilis, Aeromonas hydrophilia, Shigella spp. and Streptococcus spp. The bacterial diversity reported in this study had some similarities to previous reports by Anyanwu and Okoli (2012) who reported the presence of Enterobacter species, Alcaligenes species, E. coli, Proteus species, Klebsiella spp., P. aeruginosa, Acinetobacter spp., S. aureus and Bacillus spp. in different water supplies at Nsukka, Nigeria. Also, Umeh et al. (2020) reported the prevalence of Klebsiella pneumoniae, Acinetobacter calcoaceticus, E. coli, S. aureus, Vibrio species, Pseudomonas spp., B. subtilis, Shigella flexneri, Salmonella Typhi in selected fish ponds in Anambra State, Nigeria.
Based on the frequency rate of isolated bacteria, Acinetobacter spp. was found to be the most predominant across the four sites, constituting 50.52% of the total bacterial community. The high prevalence of this species is most likely linked to the observed discharge of abattoir effluents into the Idah River, which was also reported in the study of Tsai et al. (2018). This is also in agreement with the report of Okechi et al. (2020) who found that Acinetobacter was the most predominant bacteria in the sediment area of Otamiri River, constituting about 42.10%. In Northeastern China, Zhao et al. (2014) found that the genus Acinetobacter was also the most prominent group of isolates in zinc and arsenic polluted rivers. Species of the genera Acinetobacter are ubiquitous in a wide range of ecological areas like water, soil, sludge, and wastewater (Hamouda et al., 2011). However, some scientific reports argued that the Acinetobacter genus is a nosocomial pathogen and the possibility of it thriving in natural environments is low (Peleg et al., 2008). It would also seem that they are resilient, as documented by few reports, which showed that they can survive in various environments containing low amounts of nutritious components, as well as their resistance to adverse environmental conditions (Gospodarek and Ziolkowski, 2000). Strains of this organism are commonly known to harbor antibiotic resistance genes and they are regarded as emerging opportunistic pathogens of fish farmed in Poland (Kozi?ska et al., 2014). There might be a possibility of this occurring in Idah River, if precautionary measures are not taken.
The isolated Gram-positive bacteria, S. aureus, were also widespread across the river with a total percentage of 18.56%. The daily routine of washing, sand packing and fishing activities observed at the riverbank might account for its abundance at the docking sites. At the midstream and confluence areas of the river, its prevalence was relatively low. S. aureus is known to be commensal in the mucosa of mammals and birds. However, they can also be opportunistic pathogens (Quinn et al., 2004). The risk of S. aureus infections is premised on the possibility of its resistance to beta-lactamase antibiotics, including penicillin and thus, its presence in water calls for public health concern. Streptococcus spp. was found only in the midstream area of the river. This organism has been associated with illnesses such as pneumonia and upper respiratory tract infections. Bacillus spp. were identified across all sample sites. Bacillus spp. can occupy a wide range of ecological niches and its spores are quite ubiquitous (Nicholson, 2004). In particular, B. subtilis found in the docking and midstream area could have the potential to be used as probiotic additives in aquaculture, as reported by Guo et al. (2016).
The main indicator of faecal pollution, E. coli, was found in all sites, except for the midstream area of the water. These bacterial isolates were discovered from both docking areas, which is suggestive of sewage pollution and abbatoir effluents observed at the Idah riverbank. Contamination of the river by other members of the Enterobacteriaceae family seen in this study – Shigella spp., A. hydrophilia, P. aeruginosa and Klebsiella spp. could be related to a combination of direct faecal contamination and agricultural run-off at the docking sites. These Gram-negative bacteria are typically responsible for some waterborne diseases, including Shigellosis, typhoid, dysentery, diarrhea, as well as urinary tract infections, and they have been implicated in high mortality rate across the world (WHO, 2011). Pseudomonas spp. has been isolated from fishes in contaminated rivers. When consumed raw or insufficiently processed, such fishes could serve as a vector for the transmission of pathogens to humans (Jeyasekaran et al., 2006). Generally, the prevalence of Gram-negative bacteria in this river could be as a result of faecal contamination and other human interference, which could result in the proliferation of pathogenic organisms in fish, affecting human consumers (Kay et al., 2008). Their presence is most likely linked to the washing activities in the area, as well as the agricultural/seepage runoffs entering the river (Abulreesh, 2012). This implies a significant health risk for humans consuming this water (Franciska et al., 2005).
The mycological analysis in this study showed the presence of A. niger, A. flavus, Penicillium spp., Fusarium spp., Cladosporium spp., Trichoderma spp. and Curvularia spp. in Idah River. This agrees with the observations of Ifi et al. (2019), who reported Candida species, Fusarium spp., A. flavus, Penicillium spp., A. niger and Mucor species in Okokpon River, Edo State. Furthermore, Agbabiaka and Oyeyiola (2012) documented the presence of Curvularia, Aspergillus, Penicillium, Saccharomyces, Cladosporium, Geotrichum, Trichoderma, Mucor, Rhizopus, Fusarium and Mortierella (fungi) as sediment contaminants of Foma River, Ita-Nmo, Ilorin.
The abundance of fungi, in the docking areas of Idah River, is likely linked to its high sulphate concentration, thus constituting an indicator of water pollution from anthropogenic and agricultural origins. Pietryczuk et al. (2018) reported a similar observation in their study of fungi diversity in selected rivers. Fungal population increases with pollution and this might be connected to the high number of fungal contaminants observed at the docking areas of the river.
Amongst the various fungi species isolated in this river samples, Fusarium spp. was the most predominant, constituting about 39.13%. Fusarium spp. are plant pathogens or rhizosphere fungi, and their presence in Idah River could be attributed to agricultural activities occurring around this river. This is quite similar to the observation of Sharma and Tiwari (2015) who reported Fusarium spp. as one of the most abundant soil fungi isolated from the Shivnath River and it is known to cause several superficial infections. A. niger and A. flavus found in this river are known to be among the main agents of food spoilage but it is co-related to a range of infections such as Aspergillosis, leading to respiratory infections. Refai et al. (2010) also confirmed Aspergillus as pathogenic fungi of freshwater fishes. Trichoderma spp. was found only in DPA and DPB, which had the most observed anthropogenic activities. It is one of the beneficial fungi in the environment that can serve as a bio-fungicide against various fungal pathogens (Schuster and Schmoll, 2010). It was suggested that it might have the potential to also control infectious diseases in aquaculture (Citarasu et al., 2012). Penicillium spp. was prevalent only in the two docking points, and the genus was reported to produce mycotoxins (Pohland and Wood, 1997). Contamination of fish with these mycotoxins could accumulate in their tissues, which could invariably affect humans when consumed. The processing of fish does not necessarily eliminate the presence of mycotoxins in their tissues. Cladosporium and Curvularia spp. isolated from the docking points in the river are predominantly saprophytic in nature, and they are known to colonize and parasitize aquatic plants.
In this study, physicochemical and microbial (bacteriological and mycological) profiling of Idah River were assessed from four designated sampling points. Although, thirteen physicochemical parameters were within the safe limits as indicated by the WHO (2011) and SON (2007) standards, parameters indicative of pollution were obtained. They included high total heterotrophic bacterial counts, presence of pathogenic microbes and toxic levels of certain physicochemical factors, such as arsenic, selenium, turbidity and nickel, when compared with standards. Such extreme values derived from physicochemical analysis might be connected to anthropogenic activities of locals of the community, resulting in inorganic pollution. In addition, the various fungi and bacterial species found in the river raises health concerns, with regards to direct consumption of water and ingestion of food (such as fishes and aquatic plants) sourced from the water body. It is therefore recommended that programs and policies, such as continuous monitoring and public health awareness programs are set in place to enlighten locals of the community, about the dangers of unsuitable discharge of animal, human and inorganic wastes into the river. This could in turn, prevent the river from being an environmental reservoir of antibiotic-resistant pathogens, hence preventing public disease outbreaks.
This study provides the first report about physicochemical properties and microbial diversity (using culture-based techniques) inherent in the Idah River. However, additional microbial analysis, such as metagenomic studies, could be used to reveal a deeper microbial structure inherent in the river, which culture-dependent methodology might not have captured.
The authors have not declared any conflict of interests.
We would like to thank Mrs. Roseline Ideh of Bio entrepreneurship and Extension Services Department, National Biotechnology Development Agency (NABDA), Abuja, for her technical support. We also appreciate Ms. Halima Idris of the National Space Research and Development Agency (NASRDA), Abuja, for assisting with the GIS map of the sample sites in Idah River.
REFERENCES
|
Abulreesh HH (2012). Salmonellae in the environment. In: Annous, B, Gurtler, JB (Eds.), Salmonella-Distribution, Adaptation, Control Measures and Molecular Technologies. InTech. pp. 19-50.
|
|
|
|
Adedire OM, Atere A, Ogundipe WF, Farinu AO (2021). Effects of Direct and Indirect Sunlight on Polythene Packs, Sensory, Microbial and Chemical Properties of Sachet Water. Journal of Advances in Biology and Biotechnology pp. 25-34.
Crossref
|
|
|
|
|
Adekunle IM, Adetunji MT, Gbadebo AM, Banjoko OB (2007). Assessment of groundwater quality in a typical rural settlement in Southwest Nigeria. International Journal of Environmental Research and Public Health 4(4):307-318.
Crossref
|
|
|
|
|
Ademorati MA (1996). Standard Methods for water and effluent analysis. Foludex Press Ltd, Ibadan. 1st ed. pp. 80-83.
|
|
|
|
|
Adesakin TA, Oyewale AT, Bayero U, Mohammed AN, Aduwo IA, Ahmed PZ, Abubakar ND, Barje IB (2020). Assessment of bacteriological quality and physico-chemical parameters of domestic water sources in Samaru community, Zaria, Northwest Nigeria. Heliyon 1-14:e04773.
Crossref
|
|
|
|
|
Adeyemi SO (2010). Food and feeding habits of Synodontis resupinatus (Boulenger, 1904) at Idah area of River Niger, Kogi state, Nigeria. Animal Research International 7(3):1281-1286.
|
|
|
|
|
Agbabiaka TO, Oyeyiola GP (2012). Microbial and physicochemical assessment of Foma River, Ita-Nmo, Ilorin, Nigeria: an important source of domestic water in Ilorin Metropolis. International Journal of Plant, Animal and Environmental Sciences, 2(1):209-216.
|
|
|
|
|
Akaniwor JO, Anoksike E, Akaniuk JO (2007). Indomine industrial effluent discharge on microbial properties of new Calabar River. Scientific Research and Essays 2(1):001-005.
|
|
|
|
|
Ananthakrishnan S, Loganathan K, Ahamed AJ (2012). Study on ground water quality and its suitability or drinking purpose in alathur block-Perambalur district. Archives of Applied Science Research 4(3):1332-1338.
|
|
|
|
|
Anyanwu CU, Okoli EN (2012). Evaluation of the bacteriological and physicochemical quality of water supplies in Nsukka, Southeast, Nigeria. African Journal of Biotechnology 11(48):10868-10873.
Crossref
|
|
|
|
|
American Public Health Association (APHA) (2005). Standard methods for examination of water and wastewater (20th edn.) Washington P 1220.
|
|
|
|
|
Ayedun H, Taiwo AM, Umar BF, Oseni OA, Oderinde AA (2011). Potential groundwater contamination by toxic metals in Ifo, Southwest Nigeria. Indian Journal of Science and Technology 4(7):820-823.
Crossref
|
|
|
|
|
Bouzid M (2016). Waterborne diseases and climate change: Impact and Implications. Examining the Role of Environmental Change on Emerging Infectious Diseases and Pandemics. IGI Global. pp. 89-108.
Crossref
|
|
|
|
|
Cheesebrough M (2000). Medical Laboratory Manual for Tropical Countries. Butter worth and Co. Ltd., UK pp. 21-32.
|
|
|
|
|
Citarasu T (2012). Natural antimicrobial compounds for use in aquaculture. In: Austin B., editor. Infectious Disease in Aquaculture. Woodhead Publishing; Cambridge, UK pp. 419-456.
Crossref
|
|
|
|
|
Environmental Pollution Centers (2018). Water Pollution Causes. Available online:
View (accessed on 19 April 2018).
|
|
|
|
|
Feng BW, Li XR, Wang JH, Hu ZY, Meng H, Xiang LY, Quan ZX (2009). Bacterial diversity of water and sediment in the Changjiang estuary and coastal area of the East China Sea. FEMS Microbiology Ecology 70(2):236-248.
Crossref
|
|
|
|
|
Fernandez-Martinez A, Charlet L (2009). Selenium environmental cycling and bioavailability. A structural chemist point of view. Reviews in Environmental Science and Biotechnology 8(1):81-110.
Crossref
|
|
|
|
|
Franciska MS, Marcel D, Rinald LH (2005). Escherichia coli 0157: H7 in drinking water from private supplies. Journal of Netherlands Water Resource 39:4485-4493.
Crossref
|
|
|
|
|
Gerhardt P, Murray EGR, Wood AW, Krieg RN (1994). Methods for General and Molecular Bacteriology. ASM Press, Washington DC. pp. 340-791.
|
|
|
|
|
Gospodarek E, Zió?kowski G (2000). Antibiotic-resistant strains of Acinetobacter baumannii occurring in Poland. Epidemiologic Reviews 54:88-96.
|
|
|
|
|
Guo X, Chen DD, Peng KS (2016). Identification and characterization of Bacillus subtilis from grass carp (Ctenopharynodon idellus) for use as probiotic additives in aquatic feed. Fish and Shellfish Immunology 52:74-84.
Crossref
|
|
|
|
|
Hageskal G, Knutsen AK, Gaustad P, de Hoog GS, Skaar I (2006). Diversity and significance of mold species in Norwegian drinking water. Applied Environmental Microbiology 72(12):7586-7593.
Crossref
|
|
|
|
|
Hammer JM, Hammer JMJr (2002). Water and wastewater technology (New Delhi, Prentice-Hall).
|
|
|
|
|
Hamouda A, Findlay J, Al Hassan L, Amyes SG (2011). Epidemiology of Acinetobacter baumannii of animal origin. Journal of Antimicrobial Agents 38(4):314-318.
Crossref
|
|
|
|
|
Health Canada (2012). Guidelines for Canadian drinking water quality: Guideline technical document-Turbidity. Water and Air Quality Bureau, Healthy Environments and Consumer Safety Branch; Health Canada: Ottawa, ON, Canada.
|
|
|
|
|
Ifi F, Enerijiofi K, Ekhaise FO (2019). Investigation of the physicochemical and bacteriological qualities of Okokpon River, Edo State, Nigeria for its portability status. FUDMA Journal of Sciences 3(2):185-194.
|
|
|
|
|
Igbinosa EO, Uyi OO, Odjadjare EE, Ajuzie CU, Orhue PO, Adewole EM (2012). Assessment of physicochemical qualities, heavy metal concentrations and bacterial pathogens in Shanomi Creek in the Niger Delta, Nigeria. African Journal of Environmental Science and Technology 6:419-424.
Crossref
|
|
|
|
|
Ikhuoriah SO, Oronsaye CG, Adebanjo IA (2016). Assessment of some physical characteristics and some heavy metals of River Ossiomo, Ologbo, a tributary of the Benin River, southern Nigeria. Journal of Applied Science and Environmental Management 20(2):472-481.
Crossref
|
|
|
|
|
Jeyasekaran G, Ganesan P, Anandaraj R, Jeya Shakila R, Sukumar, D (2006). Quantitative and qualitative studies on the bacteriological quality of Indian white shrimp (Penaeus indicus) stored in dry ice. Journal of Food Microbiology 23(6):526-533.
Crossref
|
|
|
|
|
Katsoyiannis IA, Zouboulis AI, Jekel M (2004). Kinetics of bacterial As (III) oxidation and subsequent As (V) removal by sorption onto biogenic manganese oxide during groundwater treatment. Industrial and Engineering Chemistry Research 43(2):486-493.
Crossref
|
|
|
|
|
Kay D, Crowther J, Stapleton CM, Wyer MD, Ewtrell LF, Edwards A, Francis CA, McDonald AT, Watkinson J (2008). Fecal indicator organism concentrations in sewage and treated effluents. Water Resources 42:442-445.
Crossref
|
|
|
|
|
Kavka GG, Berger B, Hoch BM, Herndl, GJ (1996). Assessment of microbiological water quality in the Austrian section of the river Danube. Archiv fur Hydrobiologie, Supplementband. Larger Rivers. 113(10): 79-86.
Crossref
|
|
|
|
|
Kidu M, Abraha G, Hadera A, Yirgaalem W (2015). Assessment of physico-chemical parameters of Tsaeda-Agam River in Mekelle City, tigray, Ethiopia. Bulletin of the Chemical Society of Ethiopia 29(3):377.
Crossref
|
|
|
|
|
Kozi?ska A, Paždzior E, P?kala A, Niemczuk W (2014). Acinetobacter johnsonii and Acinetobacter iwoffii the emerging fish pathogens. Bulletin of the Veterinary Institute in Pulawy 58(2):193-199.
Crossref
|
|
|
|
|
Kuz'mina VU, Ushakova NY (2007). Metal assessment of fish ponds. Journal of Biomedical Life Sciences 47(6):837-846.
|
|
|
|
|
Leon CG, Moraga R, Valenzuela C, Gugliandolo C, Lo Giudice A, Papale M (2018). Effect of the natural arsenic gradient on the diversity and arsenic resistance of bacterial communities of the sediments of Camarones River (Atacama Desert, Chile). PLoS ONE 13(5):e0195080.
Crossref
|
|
|
|
|
Marchesi J (2017). Unlocking the Microbiome. Microbiology Society, Charles Darwin House, London. PDF file. 11 July, 2019.
|
|
|
|
|
Miller RL, Bradford WL, Peters NE (1988). Specific Conductance: Theoretical Considerations and Application to Analytical Quality Control. In U.S. Geological Survey Water-Supply Paper. Retrieved from
View
|
|
|
|
|
Murphy S (2005). General information on Dissolved Oxygen Basin Project, city of Boulder.
|
|
|
|
|
National Population Census (NPC) (2006). Federal Republic of Nigeria Official Gazette No.2 Abuja. 96 B. 25 Federal Government Printer.
|
|
|
|
|
Nicholson WL (2004). Ubiquity, longevity, and ecological roles of Bacillus spores. In Bacterial spore formers: Probiotics and emerging applications, In: E. Ricca, A.O. Henriques, and S.M. Cutting, eds. (Norflok, Horison Bioscience) pp. 1-15.
|
|
|
|
|
Nwoko CIA, Ukiwe LN, Egereonu UU, Ukachukwu SN (2015). Assessment of seasonal physicochemical parameters of Oguta Lake, Nigeria. Journal of Advances in Chemistry 11(7):3759-3764.
Crossref
|
|
|
|
|
Okechi RN, Chukwura EI (2020). Physicochemical and Bacteriological Qualities of Otamiri River Water and Sediment in South Eastern Nigeria. Frontiers in Environmental Microbiology 6(2):18-26.
Crossref
|
|
|
|
|
Okereke IJ (2014). An assessment of the physicochemical parameters of Ihuku River. IOSR Journal of Environmental Science Toxicology and Food Technology 8(1):27-30.
Crossref
|
|
|
|
|
Ologbosere OA, Aluyi HSA, Ogofure AG, Beshiru A, Omeje FI (2016). Physico-chemical and microbiological profile of bacterial and fungal isolates of Ikpoba River in Benin City: Public health implications.
Crossref
|
|
|
|
|
African Journal of Environmental Science and Technology 10(3):67-76.
|
|
|
|
|
Oremland RS, Stolz JF (2003). The ecology of arsenic. Science 300(5621):93-94.
Crossref
|
|
|
|
|
Peleg AY, Seifert H, Paterson DL (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clinical Microbiology Reviews 21(3):538-582.
Crossref
|
|
|
|
|
Pietryczuk A, Cudowski A, Hauschild T (2018). Abundance and Species Diversity of Fungi in Rivers with Various Contaminations. Current Microbiology 75(5):630-638.
Crossref
|
|
|
|
|
Premlata V (2009). Multivariant analysis of drinking water quality parameters of Lake Pichhola in Udaipur, India. Biological Forum International Journal 1(2):97-102.
|
|
|
|
|
Pohland AE, Wood GE (1997). Occurrence of Mycotoxins in Food. In: Mycotoxins in Food, Krogh, P(Ed.). Academic Press, London, UK. pp. 35-64.
|
|
|
|
|
Refai MK, Mohammed LA, Kenawy AM, Shimaa El-SMA (2010). The Assessment of Mycotic Settlement of Freshwater Fishes in Egypt. Journal of American Science 6(11):823-831.
|
|
|
|
|
Quinn PJ, Carter ME, Markey B, Garter GR (2004). Clinical Veterinary Microbiology, USA. Mosby an imprint of Elsevier Limited. pp. 43-49.
|
|
|
|
|
Schuster A, Schmoll M (2010). Biology and Biotechnology of Trichoderma. Applied Microbiology and Biotechnology 87(3):787-799.
Crossref
|
|
|
|
|
Shaji C, Nimi H, Bindu L (2009). Water quality assessment of open wells in and around Chavara industrial area, Quilon, Kerala. Journal of Environmental Biology 30(5):701-704.
|
|
|
|
|
Sharma MS, Tiwari KL (2015). Isolation and identification of soil fungi from mahamera aniket, shivnath river durg. IOSR Journal of Environmental Science, Toxicology and Food Technology 1(5):24-26.
|
|
|
|
|
Shittu OB, Olaitan JO, Amusa TS (2008). Physico-chemical and bacteriological analyses of water used for drinking and swimming purposes in Abeokuta, Nigeria. African Journal of Biomedical Research 11(3):285-290.
Crossref
|
|
|
|
|
Standard Organization of Nigeria (SON) (2007). Nigerian Standard for Drinking Water Quality, SON, Abuja. Nigeria.
|
|
|
|
|
Taiwo AA, Abayomi TO, Umar B, Abubakar NM, Iduwo AA, Ahmed PZ, Abubakar ND, Ibrahim BB (2020). Assessment of bacteriological quality and physico-chemical parameters of domestic water sources in Samaru community, Zaria, Northwest Nigeria, Heliyon 6(8) 04773-8440.
Crossref
|
|
|
|
|
Thi TGL, Suthipong S, Kyoung-Woong, K (2009). Arsenic and other trace elements contamination in groundwater and a risk assessment study for the residents in the Kandal Province of Cambodia. Environment International 35(3):455-460.
Crossref
|
|
|
|
|
Tsai HC, Chou MY, Shih YJ, Huang TY, Yang PY, Chiu YC, Chen JS, Hsu BM (2018). Distribution and genotyping of aquatic Acinetobacter baumanii strains isolated from the Puzi River and its tributaries near areas of livestock farming. Water 10:1374.
Crossref
|
|
|
|
|
Umeh OR, Chukwura EI, Ibo EM (2020). Physicochemical, bacteriological and parasitological examination of selected fish pond water samples in Awka and its environment, Anambra State, Nigeria. Journal of Advances in Microbiology pp. 27-48.
Crossref
|
|
|
|
|
U.S. Environmental Protection Agency (EPA) (2016). Six-Year Review 3 Technical Support Document for Microbial Contaminant Regulations. Available online at: View
|
|
|
|
|
Venkatasubramani R, Meenambal T (2007). Study on subsurface water quality in Mettupalayam taluk of Coimbatore district, Tamil Nadu. Nature Environment and Pollution Technology 6(2):307-310.
|
|
|
|
|
World Health Organization (WHO) (2011). Guidelines for Drinking-Water Quality, Fourth 565 edition. WHO Library Cataloguing-in-Publication, Switzerland.
|
|
|
|
|
World Health Organization (WHO) (2014). The United Nations International Children's Emergency Fund (UNICEF). Joint Monitoring Programme for Water Supply and Sanitation. In Progress on Drinking Water and Sanitation: 2014 Update; UNICEF: New York, NY, USA.
|
|
|
|
|
Zhao J, Zhao X, Chao L, Zhang W, You T, Zhang J (2014). Diversity change of microbial communities responding to zinc and arsenic pollution in a river of northeastern China. Journal of Zhejiang University-Science B Biomedicine and Biotechnology 15(7):670-680.
Crossref
|
|