Full Length Research Paper
ABSTRACT
Antibiotics are the most successful form of therapeutics developed for the treatment of disease caused by bacteria. The study aimed to assess the prevalence of Escherichia coli and multidrug resistant pattern from environmental sources in Port Harcourt, Rivers State, Nigeria. Forty samples were collected from environmental sources including poultry litter, soil, waste water and cloaca. All samples were inoculated onto prepared Eosin Methylene blue plates and incubated for 24 h at 37°C. Colonies were sub cultured onto sterile nutrient agar plates. Pure isolates were identified using standard microbiological methods. Antibiotic susceptibility was carried out on identified E. coli. The study showed that from the samples poultry had 15 (37.5%) E. coli, soil 11 (27.5%), waste water 9 (22.5%) and cloaca 5 (12.5%) E. coli. However, the highest number of E-coli was observed in poultry source and least in cloaca. The results also revealed that the number of E. coli from poultry were 7 (46.7%), 5 (33.3%), 2 (13.3%) and 1 (6.7%), soil 6 (54.5%), 1 (9.1%), 3 (27.3%) and 1 (9.1%), waste water 2 (22.2%), 2 (22.2%), 2 (22.2%), 2 (22.2%) and 1 (11.1%) and cloaca 2 (40.0%) and 3 (60.0%), respectively. E.-coli were susceptible and resistant to classes of antibiotic including Ceftazidime, Cefuroxime, Gentamicin, Cefxime, Ofloxacin, Augmentin, Nitrofurantoin and Ciprofloxacin. Hence, the study s amongst others that to prevent further emergence and spread of resistant strains in E-coli, rational use of antibiotics and regular monitoring of antimicrobial resistance patterns are essential and mandatory.
Key words: Antibiogram, Escherichia coli, environment, multidrug, resistance.
INTRODUCTION
Escherichia coli are Gram negative pathogen with a global distribution rate. It can be isolated from environmental, clinical, and animal sources. Certain strains of E. coli cause most clinical and environmental mediated diseases. Antibiotic resistance has become a worldwide concern due to the emergence of antibiotic-resistant bacteria which limits the clinical use of antibiotics. Antibiotic resistance increases the prevalence of resistant bacteria in both clinical and environmental sources thus rendering available antibiotics ineffective for therapeutic purposes (Ajuga et al., 2021; Odonkor and Kenned, 2018; Agbagwa and Jirigwa, 2015). Sommer et al. (2017) reported that antibiotic-resistant genes responsible for resistance to a wide variety of antibiotics have been identified in a large range of environments including drinking water, waste water, soil and cloaca, etc., in both developed and developing countries. The main risk for public health is that resistance genes are transferred from environmental bacteria to human pathogens. The potential of the environment to transport microbial pathogens to a greater number of people, causing subsequent illness, is well documented in countries at all levels of economic development. Furthermore, the availability of safe environment is an indispensable feature for preventing epidemic disease and improving the quality of life. Hence, the World Health Organization reported that 80% of all diseases are attributed to unsafe environment. This is to say that developing countries in particular, are plagued with water-related diseases such as diarrhoea which account for 10% of the disease burden in such countries (Ellis and Schoenberger, 2017). Availability of safe environment is a key factor underpinning public health and development of any nation. Environmental sources that may harbour microorganisms include surface water such as lakes, streams, rivers, ponds and underground water such as springs, wells, borehole, soil and animals houses (Oluyege et al., 2009). Lyimo et al. (2016) reported that 748 million, mostly poor and marginalized people still lack access to quality drinking water and safe environment. Of these, almost a quarter (173 million) rely on untreated surface water on a daily basis and over 90% live in rural areas as faecal waste from people and animals is a major source for pollution, particularly in low-income countries.
In 2012, reports had it that approximately 1 billion people in the world did not have access to toilet facilities and instead used open and unsanitary places for defecation especially water bodies. These communities also lack proper water supplies and depend heavily on untreated surface or shallow, unprotected water sources for consumption. Strains of E. coli that are pathogenic to both humans and animals are capable of causing disease ranging from self-limiting diarrhoea to life-threatening haemolytic-uremic syndrome and haemorrhagic colitis. However, studies have revealed non-conformity of many water sources in Nigeria to World Health Organization (WHO) standard which has led to faecal contamination of water sources which can extend to other sources (Oluyege et al., 2009). The emergence and dissemination of antimicrobial-resistant (AMR) bacteria is considered the third-largest threat to global public health in the 21st century which reduces the effectiveness of antibiotic treatment and thus leads to increased morbidity, mortality, and healthcare expenditures (WHO, 2014). Hence, the E. coli found in people and animals is considered a potential reservoir for AMR genes and these genetic traits can be transferred to or to other bacteria found in people, animals, and in the environment (Katakweba et al., 2018). The study intends to assess the prevalence of E. coli and multidrug resistant pattern from environmental sources in Port Harcourt, Rivers State, Nigeria to provide and guide concerted policies for necessary interventions.
MATERIALS AND METHODS
Study area
The research was carried out at the Medical Laboratory of the Department of Microbiology in the University of Port Harcourt which is located at Choba, Rivers State, Nigeria.
Sample collection
Forty samples were collected from environmental sources including poultry, soil, waste water and cloaca. 15 samples were from poultry, soil (11), waste water (9), cloaca and (5). All samples were preserved in cold boxes, transported to the Medical Laboratory of the Department of Microbiology in the University of Port Harcourt within 4 h and maintained at 4°C until use.
Isolation and identification of E. coli
All environmental samples (poultry, soil, waste water soil and cloaca) were inoculated on prepared Eosin Methylene Blue (EMB) agar plates and incubated for 24 h at 37°C. The colonies on the plates were sub cultured onto nutrient agar plates (Oxiod) to obtain pure colony. Pure colonies were stored and subjected to Gram staining selected biochemical test such as: citrate test, indole test, oxidase test, triple sugar iron agar test, methyl red and Voges-Proskuer test for identification (Cheesbrough, 2006). They were further confirmed using E. coli specific 16s rRNA gene fragment of Ec16 primer pairs (F 5’-GACCTCGGTTAGTTCACAGA-3’ and R 5’-CACACGCTGACGCTGACCA-3’) (Islam et al., 2016). The reaction mixture was prepared by the addition of 3 µl of E. coli DNA, 10 µl PCR master mix, 1 µl of each of the two primers and 6 µl of nuclease free water. The primers have an annealing temperature of 55°C and result in a product with base pair of 588 bp (Islam et al., 2016).
Antibiotic susceptibility testing
Antibiotic susceptibility testing was carried out on identified isolates by the disc diffusion method (CLSI, 2014). In brief, isolates were inoculated on sterile nutrient broth for 16 to 18 h of incubation at 37°C. Inoculum size was adjusted 0.5 McFarland standards and swabbed onto Muller-Hinton agar. Antibiotic disc was placed and plates were incubated for 24 h at 37°C. The zone of inhibition was measured to the nearest millimetre and all bacterial isolates were classified as sensitive, intermediate, and resistant.
RESULTS
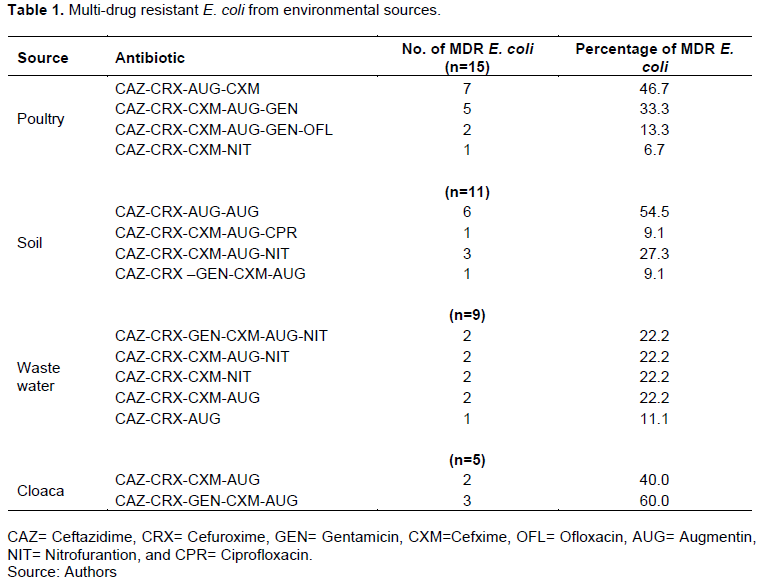
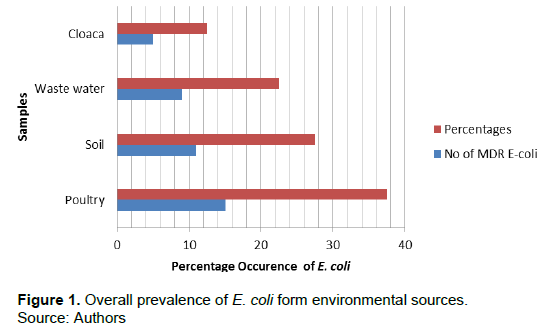
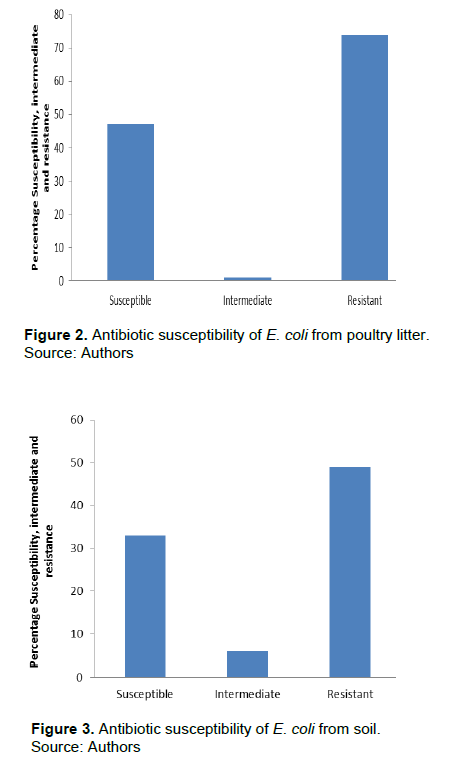
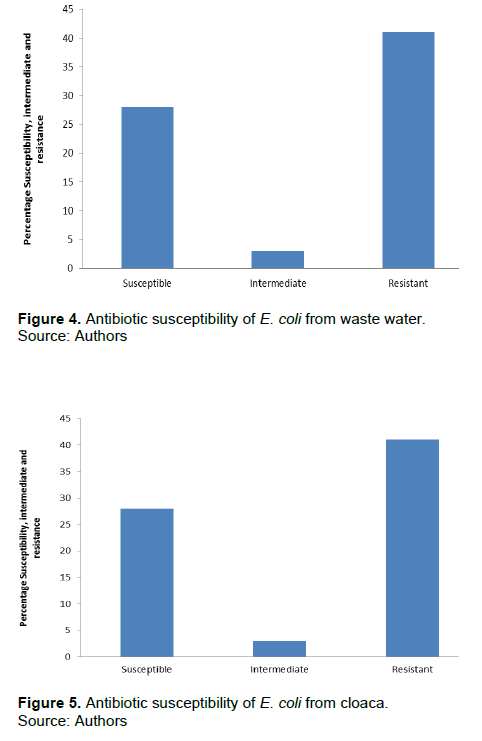
Of the fifty samples collected from various sources, poultry had 15 (37.5%) E. coli samples, soil 11 (27.5%), waste water 9 (22.5%) and cloaca 5 (12.5%) E.coli. However, the highest number of E. coli was observed in poultry source and least in cloaca sample. Detailed result of the overall prevalence of E.coli is presented in Figure 1. Table 1 shows that the number of MDR E. coli from poultry was 7 (46.7%), 5 (33.3%), 2 (13.3%) and 1 (6.7%), soil 6 (54.5%), 1 (9.1%), 3 (27.3%) and 1 (9.1%), waste water 2 (22.2%), 2 (22.2%), 2 (22.2%), 2 (22.2%) and 1 (11.1%) and cloaca 2 (40.0%) and 3 (60.0%), respectively. The identified 40 E. coli were subjected to antibiotic susceptibility testing. Results obtained showed that E. coli from poultry was 47% susceptible, 1% intermediate, and 74% resistant to antibiotic susceptibility test (Figure 2). E. coli from soil (Figure 3) was 33% susceptible, 6% intermediate and 49% resistant to the antibiotic tested. E. coli from waste water (Figure 4) was 28% susceptible, 3% intermediate, and 41% resistant to antibiotic susceptibility test and Figure 5 shows that E. coli from cloaca was 16% susceptible, 1% intermediate, and 23% tested antibiotics.
DISCUSSION
The aim of the study was to assess the prevalence of E. coli and multidrug resistant pattern from environmental sources. The finding of the study showed that E. coli were isolated from poultry, soil, waste water and cloaca. Fifteen numbers of E. coli samples were isolated poultry, soil 11, waste water 9, and cloaca 5. Detailed results are as shown Figure 1 (Overall prevalence of E. coli). Isolates were identified by standard microbiological methods. However, colonial morphology for identification is presented. The results showed that from the samples collected from various sources, poultry had 15 (37.5%) E. coli samples, soil 11 (27.5%), waste water 9 (22.5%) and cloaca 5 (12.5%) E. coli, this shows the presence of multi-drug resistant E. coli in the various samples. The finding of this study confirms that of Galindo-Mendez (2020), Singh et al. (2020) in Indian whose studies reported the prevalence of antibiotic resistant genes among multi-drug resistant E. coli. However, these studies were sampled in human faeces and at least two antibiotic classes were detected. The finding of this study is in conformity with that of Rubab and Oh (2021), Jahantigh et al. (2020) whose studies discovered the presence of multi-drug resistant in E. coli. However, most of these studies were done among STEC isolates and lesions in broiler chickens with gentamicin being the most resistant. By implication, these results indicated that there is high level of the prevalence of multi-drug resistant E. coli both in the studied area and other studies as confirmed by Adesoji et al. (2015) and the present study. The present study disagrees with the study carried out by some researchers where the resistant level was higher than the present study. This difference observed could be attributed to the environmental factors, the strain, samples source and other factors (Karami et al., 2006; Xi et al., 2009; Coleman et al., 2012; Chen et al., 2017; Sanganyado and Gwenzi, 2019; Praveenkumarreddy et al., 2020). Multidrug resistant E. coli is currently on the increase and more prevalent in developing countries where antibiotic are used indiscriminately in agriculture, veterinary and medicine. Antibiotics are used in agriculture and animals without proper investigation and policies to guide the use of antibiotics. This can be a major avenue for the transfer of antibiotic resistant bacteria to humans via contaminated environmental sources (Ukah et al., 2018).
CONCLUSION
This study shows the presence of multi-drug resistant E. coli with most showing susceptible and resistance to classes of antibiotic including Ceftazidime, Cefuroxime, Gentamicin, Cefxime, Ofloxacin, Augmentin, Nitrofurantoin and Ciprofloxacin. Hence, to prevent further emergence and spread of MDR resistant E. coli, policies guiding the use of antibiotics and regular monitoring of antimicrobial resistance patterns should be put in place to prevent the transfer of resistant bacteria from one source to another.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
REFERENCES
|
Agbagwa OE, Jirigwa CE (2015). Antibiotics resistance and plasmid profile of Staphylococcus aureus from wound swabs in Port Harcourt Nigeria. Current Research in Bacteriology 8(3):70-76. |
|
|
Ajuga MU, Otokunefor K, Agbagwa OE (2021). Antibiotic resistance and ESBL production in Escherichia coli from various sources in Aba metropolis, Nigeria Bulletin National Research Center 45(1):173. |
|
|
Adesoji AT, Ogunjobi AA, Olatoye IO, Douglas DR (2015). Prevalence of tetracycline resistance genes among multi-drug resistant bacteria from selected water distribution systems in southwestern Nigeria. Annals of Clinical. Microbiology Antimicrobials 14(1):1-8. |
|
|
Cheesbrough M (2006). District Laboratory Practice in Tropical countries Part 1&2. Second edition update, Cambridge university press. |
|
|
Chen Z, Yu D, He S, Ye H, Zhang L, Wen Y, Chen S (2017). Prevalence of antibiotic-resistant Escherichia coli in drinking water sources in Hangzhou city. Frontiers Microbiology 8: 1133. Clinical and Laboratory Standards Institute (2014). Performance standards for antimicrobial susceptibility testing; fifteenth informational supplement. Clinical and Laboratory Standard Institute Wayne, Pa. M100- S15, 25:1. |
|
|
Coleman BL, Salvadori MI, McGeer AJ, Sibley KA, Neumann NF, Bondy SJ, Gutmanis IA, McEwen SA, Lavoie M, Strong D, Johnson I (2012). The role of drinking water in the transmission of antimicrobial-resistant E. coli. Epidemiology and Infection 140(4):633-642. |
|
|
Ellis H, Schoenberger E (2017). On the identification of associations between five world health organization water, sanitation and hygiene phenotypes and six predictors in low and middle-income countries. PloS One 12(1):e0170451. |
|
|
Galindo-Méndez M (2020). Antimicrobial Resistance in Escherichia coli. E. coli Infections-Importance of Early Diagnosis and Efficient Treatment pp. 1-20. |
|
|
Islam MA, Kabir SML, Seel SK (2016). Molecular Detection and Characterization of Escherichia coli isolated from Raw milk sold in different markets of Bangladesh. Bangladesh Journal of Veterinary Medicine 14(2):271-275. |
|
|
Jahantigh MK, Samadi RE, Dizaji, Salari S (2020). Antimicrobial resistance and prevalence of tetracycline resistance genes in Escherichia coli isolated from lesions of colibacillosis in broiler chickens in Sistan, Iran. BMC Veterinary Research 16:1-6. |
|
|
Karami N, Nowrouzian F, Adlerberth I, Wold AE (2006). Tetracycline resistance in Escherichia coli and persistence in the infantile colonic microbiota. Antimicrobial Agents and Chemotherapy 50(1):156-161. |
|
|
Katakweba AA, Muhairwa AP, Lupindu AM, Damborg P, Rosenkrantz JT, Minga UM, Mtambo MM, Olsen JE (2018. First report on a randomized investigation of antimicrobial resistance in fecal indicator bacteria from livestock, poultry, and humans in Tanzania. Microbial Drug Resistance 24(3):260-268. |
|
|
Lyimo B, Buza J, Subbiah M, Smith W, Call DR (2016). Comparison of antibiotic resistant Escherichia coli obtained from drinking water sources in northern Tanzania: a cross-sectional study. BMC Microbiology 16(1):1-10. |
|
|
Odonkor ST, Kennedy KA (2018). Prevalence of Multidrug-Resistant Escherichia coli Isolated from Drinking Water Sources, International Journal of Microbiology ID 7204013. |
|
|
Oluyege JO, Dada AC, Odeyemi AT (2009). Incidence of multiple antibiotic resistant Gram-negative bacteria isolated from surface and underground water sources in south western region of Nigeria. Water Science and Technology 59(10):1929-1936. |
|
|
Rubab M, Oh DH (2021). Molecular Detection of Antibiotic Resistance Genes in Shiga Toxin-Producing E. coli Isolated from Different Sources. Antibiotics 10(4):344. |
|
|
Sanganyado E, Gwenzi W (2019). Antibiotic resistance in drinking water systems: Occurrence, removal, and human health risks. Science of Total Environment 669:785-797. |
|
|
Singh BR, Singh SV (2020). Metallo-β-Lactamase and Extended-Spectrum-β-Lactamase Production by Serratia Strains. Infection and Drug Resistance 13:1295. |
|
|
Sommer MO, Munck C, Toft-Kehler R, Andersson DI (2017). Prediction of antibiotic resistance: time for a new preclinical paradigm? Nature Review Microbiology 15(11):689-696. |
|
|
Ukah UV, Glass M, Avery B, Daignault D, Mulvey MR, Reid-Smith RJ, Parmley EJ, Portt A, Boerlin P, Manges AR (2018). Risk factors for acquisition of multidrug-resistant Escherichia coli and development of community-acquired urinary tract infections. Epidemiology and Infection 146(1):46-57. |
|
|
World Health Organization (WHO) (2014). "Antimicrobial resistance: global report on surveillance 2014". WHO. Archived from the original on 15 May 2015. Retrieved 9 May 2015. |
|
|
Xi C, Zhang Y, Marrs, CF Ye W, Simon C, Foxman B, Nriagu J (2009). Prevalence of antibiotic resistance in drinking water treatment and distribution systems. Applied and Environmental Microbiology 75(17):5714-5718. |
|
|
Praveenkumarreddy Y, Akiba M, Guruge KS, Balakrishna K, Vandana KE, Kumar V (2020). Occurrence of antimicrobial-resistant Escherichia coli in sewage treatment plants of South India. Journal of Water, Sanitation and Hygiene for Development 10(1):48-55. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0