ABSTRACT
For many years, the generation of nitrates from organic sources in order to create nutrient solutions for hydroponics had proved a challenge until lately when microorganisms were introduced to perform this task. The objectives of the current study therefore were to use local microbial consortium to nitrify goat manure in water and to determine microbial diversity in the inoculated consortium. Therefore, microorganisms were sourced from garden soil and natural compost at the Sam Nujoma Marine resources Research Centre (SANUMARC) in Henties Bay Namibia to convert organic nitrogen in goat manure from Utuseb farm near Walvis Bay into nitrates. Results show that microbial consortium from the compost source produced significantly (P< 0.05) more nitrates followed by the garden-soil source, suggesting that it is necessary to add inoculum in order to generate nitrate from goat manure. The ammonia oxidising bacteria (AOB) community from the compost sample’s was dominated by uncultured ammonia-oxidising species followed by uncultured bacterium (both not identified), with the least being Nitrosomonas species. The AOB community from the garden source was dominated by uncultured bacterium, followed by uncultured ammonia-oxidising species and the least being Nitrosomonas species. NOB community from the compost source was dominated by uncultured bacterium, followed by Nitrobacter winogradskyi and Nitobacter vulgaris with the least being Nitrococcus mobilis and Nitrospira moscovensis. For the garden soil microbial source, uncultured nitrite-oxidising bacteria dominated followed by uncultured bacterium, whereas the least species were N. moscovensis and Nitrobacter alkalicus. Moreover, community composition of the compost sample was more diverse than the community from the garden sample. These results maintain that there are other unculturable yet important microbes doing the same job if not better than the known ones, in this case suggesting that there may be other local nitrite-oxidizing bacteria responsible for oxidizing ammonia other than the traditionally known Nitrobacter, Nitrospira and Nitrococcus species.
Key words: Organic hydroponics, ammonification, nitrification, ammonia-oxidizing bacteria, nitrite-oxidizing bacteria.
For many years, the generation of nitrates from organic sources in order to create nutrient solutions for hydroponics had proved a challenge. It was not until Shinohara et al. (2011) used nitrifying microorganisms to degrade organic nitrogen into nitrates, which is directly used by plants, that organic hydroponics have been made possible with follow-up studies such as that of Hu and Qi (2013) confirming the successful use of microbes in nitrification of other organic sources. Nitrification consists of two consecutive oxidation steps: nitritation and nitratation done by two main groups of bacteria (ammonia-oxidizing bacteria (AOB) and nitrite-oxidizing bacteria (NOB)). Most AOBs belong to the beta-subclass of Proteobacteria and four genera belonging to this lineage are Nitrosomonas (with Nitrosococcus mobilis), Nitrosospira, Nitrosolobus and Nitrosovibrio (Woese et al., 1984; Pommering-Röser et al., 1996). NOBs on the other hand consist of the Proteobacteria genera Nitrobacter, Nitrococcus and Nitrospina. The genus Nitrobacter has four species Nitrobacter winogradskyi, Nitrobacter hamburgensis, Nitrobacter vulgaris and Nitrobacter alkalicus (Daims, 2001). The genera, Nitrococcus contains Nitrococcus mobilis, whereas the genera Nitrospina contain Nitrospina gracilis. The genus Nitrospira consists of N. marina from ocean water and N. moscoviensis (Daims, 2001).
These microorganisms can be cultured and stored for use in water and soil media to degrade organic nitrogen (Willuweit et al., 2008). All they require is an ecosystem with favourable conditions for them to function in their specific diversity (Saijai et al., 2016). But culturing has a major disadvantage in that many nitrifying bacteria are not cultivable. It is estimated that in seawater samples at best, only 0.1% (Kogure et al., 1980), in freshwater only 0.25% (Jones, 1977), in soil samples 0.5% (Torsvik et al., 1990), and in activated sludge 15% (Kämpfer et al., 1996) of the indigenous bacteria could be cultivated. Traditionally, Nitrosomonas and Nitrobacter were thought to be the only microbes responsible for nitrification in wastewater treatment plants based on the experience that Nitrosomonas and Nitrobacter species can be isolated from every activated sludge. It was only when Nitrobacter was not detected in an aquarium, yet there was nitrification, that it was considered that there are other unculturable yet important microbes doing the same job. Therefore, microbial ecology needs cultivation-independent tools to quantify bacteria directly in environmental samples (Daims, 2001).
It has been established that under 40% of soil water filled pore space decreases abundance of AOB than over 40% (Barrena et al., 2017). This abundance is influenced by pH of soil, with acidic soils (4.0/5.4) negatively affecting abundance of AOBs, whereas more AOBs positively correlate with nitrification (Duncan et al., 2017). Furthermore, AOA may affect N cycling more in soils receiving animal manures, whereas AOB are functionally more important in chemically fertilized soils (Zhou et al., 2015). Moreover, abundances of archaeal 16S rRNA and amoA genes have been found to be positively correlated with soil nitrate, N and C contents (Rughöft et al., 2016), whereby changes to physical properties of soil determine nitrifying and ammonifying capacity and mineral nitrogen content. Maximum content of nitrate and mineral nitrogen and the biggest nitrifying capacity are at soil temperatures of 15°C, whereas temperature at -4°C have recorded the lowest abundances of NOBs (Wertz et al., 2013).
Moistening conditions optimal for developing nitrifying and ammonifying bacteria are formed at soil moisture of 20-25% (60-75% WFC) (Evdokimova et al., 2016). Functional genes have been found to be good molecular markers for studying the diversity within functional groups (Poly et al., 2008; Calvo et al., 2005). For nitrification, the amoA gene encoding ammonia mono-oxygenase is being used for microbial diversity and phylogenetic characteristics of soil and water AOB com-munities (Rotthauwe et al., 1997; Purkhold et al., 2000). In Nitrobacter, the oxidation of nitrite to nitrate is performed by the nitrite oxidoreductase (NXR), encoded by the nxr operon (Starkenburg et al., 2006) previously called nor (Kirstein and Bock, 1993). Due to ability to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world (Marco, 2011).
The need to incorporate metagenomics has been driven by the inability to culture the majority of microbes from an ecosystem and the logistics of using a myriad of media and culturing conditions to capture those that can be grown in vitro (Marchesi, 2012). The advent of next generation sequencing (NGS) technologies allow us to sequence DNA and RNA much more quickly and cheaply than the previously used Sanger sequencing due to the inexpensive production of large volumes of sequence data (Metzker, 2010). The results can thereafter be put through binning where methods such as Basic Local Alignment Search Tool (BLAST) are used to search for phylogenetic markers or similar sequences in existing public databases (Huson et al., 2007). The objectives of the current study therefore were to use local microbial consortium to nitrify goat manure in water and to determine microbial diversity in the inoculated consortium.
Study site
The current study was conducted at the Sam Nujoma Marine Resources Research Centre (SANUMARC), Henties Bay, Namibia (Figure 1).
Manure collection
Dry manure was sampled according to Lupwayi et al. (2000), whereby samples were collected from Utuseb farm in the Erongo region, where the manure was stored for at least six months. The manure was homogenized, air-dried for 1 week at 30°C, ground and sieved (< 2 mm) before use with microorganisms (Qian and Schoenau, 2001). Goat manure was chosen due to its availability at coastal areas in Namibia as compared to other forms of manure.
Source of microorganisms
The following sources were used: Sam Nujoma Marine Resources Research Centre (SANUMARC) garden soil and SANUMARC compost. SANUMARC garden soil was soil extracted from garden plots in the shaded-net garden of SANUMARC at Henties Bay. SANUMARC compost was soil collected from a compost heap at SANUMARC where grass clippings and plant leaves have been heaped for longer than 3 months. In order to determine the best source of microorganisms to mineralize organic N, samples from the above sources were investigated as treatments (Table 1). Each treatment was added to separate one-litre flasks of distilled water containing 1 g/L of goat manure. Each treatment was triplicated. The flasks were shaken (120 strokes/min) for 20 days at 25°C, and the NO3− concentrations were then determined using the Hach DR5000 spectrophotometer which uses cadmium metals to reduce nitrates in the sample to nitrite. The nitrite ion reacts in an acidic medium with sulfunic acid to form an intermediate diazonium salt. The salt couples with gentisic acid to form an amber colored solution and the results are then measured at 500 nm.

Identification of microbes
In order to understand microbial composition of the 2 inocula from the source of microorganisms experiment, identification was done. Therefore, DNA was extracted from both the garden soil and the compost from SANUMARC. The DNA extraction was performed according to the manufacturer's recommendations, based on direct cell lysis with subsequent recovery and purification of nucleic acids. Up to 250 mg of soil was added to the lysis tubes, and the samples were homogenized using a standard benchtop vortex for 20 min. The DNA was then amplified using functional genes targeting primers (NorA and AmoA) using standard PCR procedure at SANUMARC. Genomic DNA was prepared and used as a template. 305 bp NxrA genes and 490 bp AmoA genes were amplified using NorA and AmoA genes. The sequence of the amoA primers was: amo r5-′ GGGGTTTCTACTGGTGGT-3′, amoA-r 5′- CCCCTCKGGAAAGCCTTCTTC-3′ (Rotthauwe et al., 1997; Purkhold et al., 2000). The sequence for the NXR primers was: norA-f 5′ CAGACCGACGTGTGCGAAAG3′, norA-r 5′-TCYACAAGGAACGGAAGGTC-3′ (Poly et al., 2008).
The PCR mixture included 0.5 µM of each primer, 23.5 µl of nuclease free water, 25 µl master mix and 1 µl of DNA template, making up a total of 50 µl. The negative control contained nuclease free water in place of DNA template. Using the thermos-cycler, the PCR reaction was carried out as follows: NXRA- 1 cycle of pre-denaturation at 94°C for 5 min, 30 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, extension at 72°C for 45 s, final extension at 72°C for 5 min. AmoA- 1 cycle of pre-denaturation at 94°C for 5 min, annealing at 60°C for 1 min 30 s, extension at 72°C for 1 min 30 s, 42 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min 30 s, final extension at 72°C for 10 min. Amplified PCR products were visualised on 1% agarose gel with a 1kb ladder (Inqaba Biotechnology Industries, SA). Further sequencing of the PCR products was done at Inqaba Biotechnology in South Africa and the reads were merged, clustered and then a BLAST search was done against Genbank in order to identify nitrifying species from the sequence data.
Statistical analysis
Means comparisons and correlations analysis were done whereby, a one-way analysis of variance (ANOVA), followed by mean separation by Duncan’s multiple range test was used when ANOVA determined that the effects of the treatments were significant (p < 0.05 for F-test).
Yij= μ+τi + βj + γij + ϵij
Where, μ is the overall mean response, τi is the effect due to the i -th level of factor A, βj is the effect due to the j -th level of factor B and γij is the effect due to any interaction between the i -th level of A and the j -th level of B, ϵij is the error term where the error terms are independent observations. The Simpson’s diversity index was used to calculate for microbial species diversity (Atlas and Bartha, 1998):
Sáµ¢ = 1-D (∑n(n-1)/N(N-1)
Where, n is the total number of organisms of a particular species, and N is the total number of individuals of all species. Simpson's diversity index is a measure of diversity in ecology where it is used to quantify the biodiversity of a habitat taking into account the number of species present, as well as the abundance of each species.
Garden soil and compost soil were chosen as inoculum sources for the microorganisms needed for mineralization and nitrification of organic nitrogen into nitrate in water in the presence of goat manure as a nitrogen source. The pH of the garden soil was 7.8, whereas for compost soil, pH was 8.6 with low holding water capacity. The soil temperature ranged between 14.8 and 25°C, whilst soil moisture was 73% water field capacity (WFC) for garden soil and 65% WFC for compost soil. The addition of only goat manure without inoculum in water resulted in the generation of nitrate (Figure 2). In contrast, using inoculum such as nursery or compost resulted in more mineralization of organic nitrogen into nitrate was made from the addition of microbial consortiums (Figure 2).
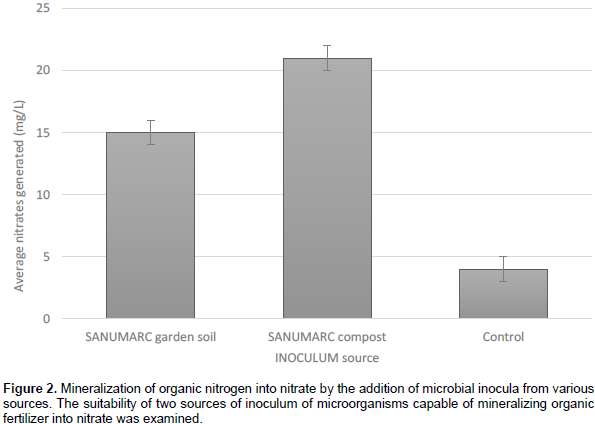
The microbial consortium from the compost source however produced significantly (P<0.05) more nitrates than the garden-soil source and the control where no inoculum was added (Figure 2). Garden soil inoculum also produced more (P<0.05). These results indicate that microorganisms added to the water from the garden soil and from compost were able to mineralize and nitrify organic nitrogen into nitrate. These results therefore suggest that it is necessary to add inoculum in order to generate nitrate from goat manure, just as it is for other organic sources such as fish-based fertiliser established by Shinohara (2011). The results are indicative of the fact that microorganisms in the inoculum can degrade organic nitrogen to nitrates but leaves the question of what is the composition of local nitrifiers within the inoculum capable of ammonification and nitrifying Therefore for the next step, all nitrifiers were identified.
Ammonia oxidizing bacteria
Figure 3 shows that from the compost sample, abundance of bacterial amoA gene was dominated by uncultured ammonia-oxidising species followed by uncultured bacterium (both not identified), with the least being Nitrosomonas species. The AOB community from the garden source was however dominated by uncultured bacterium, followed by uncultured ammonia-oxidizing species and the least being Nitrosomonas species. Moreover, the Simpson’s Diversity index of diversity showed that microbial community from the garden soil was more diverse than the microbial community from the compost sample (Table 2).
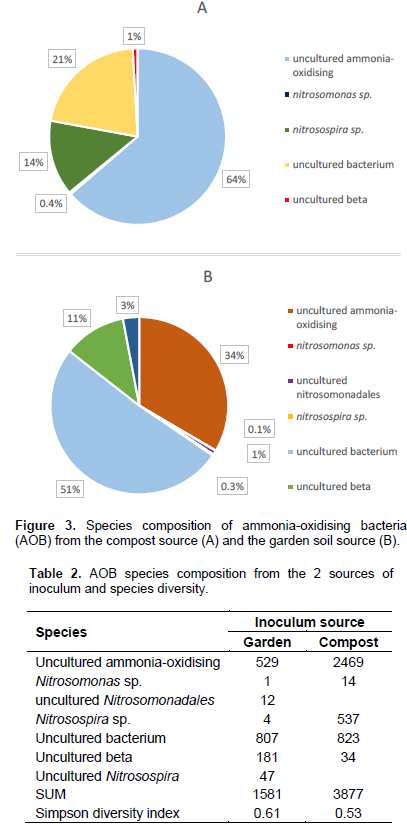
This is because community composition between the two samples differed in that more than two-thirds was dominated by a single species in the compost sample as compared to only half of the dominance given to a single species from the garden soil sample. These results concur with that of Daims (2001) who maintained that there are other unculturable yet important microbes doing the same job if not better than the known ones, in this case, suggesting that there may be other local ammonia-oxidising bacteria responsible for oxidizing ammonia other than the traditionally known Nitrosomonas and Nitrosospira species. This is considering that nitrifying bacteria picked up by the AmoA gene encoding the active site polypeptide of the ammonia monooxygenase (AmoA) in our samples are dominated by species unidentified.
Nitrite oxidizing bacteria (NOB)
Figure 4 shows that NOB community from the compost source was dominated by uncultured bacterium, followed by N. winogradskyi and N. vulgaris with the least being N. mobilis and N. moscovensis. For the garden soil microbial source, uncultured nitrite-oxidising bacteria dominated followed by uncultured bacterium, whereas the least species were N. moscovensis and N. alkalicus. Moreover, community composition of the compost sample was more diverse than the community from the garden sample (Table 3). Though community composition follows similar trend as with AOB found in this study in that the dominant nitrifying species in the samples were unidentified species, there are significant proportions of Nitrobacter species (Figure 4).
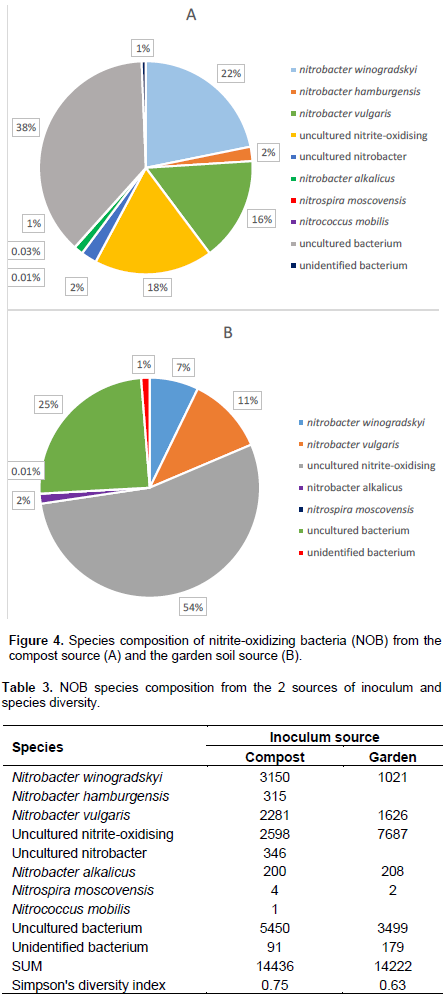
This result upholds the findings by Saijai et al. (2016), Poly et al. (2008), Wertz et al. (2012) and Winslow (1917) who established that Nitrobacter species were responsible for converting nitrite into nitrate. These results further maintain that of Daims (2001) who held that there are other unculturable yet important microbes doing the same job if not better than the known ones, in this case, suggesting that there may be other local nitrite-oxidizing bacteria responsible for oxidizing ammonia other than the traditionally known Nitrobacter, Nitrospira and Nitrococcus species.
Garden soil and compost are appropriate sources of microorganisms to generate nitrate from goat manure in water. There are other unknown local microorganisms oxidising ammonia and nitrite and the presence of these nitrifying bacteria in addition to the presence of the known ammonia-oxidizing species in the garden soil and compost samples infers their roles in the observed nitrification. Therefore, nitrifying microbial consortia that would degrade goat manure for use in organic hydroponics can be sourced from any place with similar environment (temperature, pH and soil) to Henties Bay. Further research can consider determining ammonium levels as well from nitrification of goat manure and establish the optimal rate of nitrification by a certain amount of soil containing nitrifying microbial consortia.
The authors have not declared any conflict of interests.
REFERENCES
|
Arroukatchee (2017). Map of Henties Bay.
View
|
|
|
|
Atlas R, Bartha R (1998). Microbial ecology: Fundamentals & Applications. 4th Edition. Benjamin Cummings Pubs. University of Califonia.
|
|
|
|
|
Barrena I, Menéndez S, Correa-Galeote D, Vega-Mas I, Bedmar EJ, González-Murua, C, Estavillo JM (2017). Soil water content modulates the effect of the nitrification inhibitor 3,4-dimethylpyrazole phosphate (DMPP) on nitrifying and denitrifying bacteria. Geoderma 303:1-8.
Crossref
|
|
|
|
|
Calvo L, Cortey M, Garcia-Marin JL, Garcia-Gil LJ (2005). Polygenic analysis of ammonia-oxidizing bacteria using 16S rDNA, amoA, and amoB genes. Intl. Microbiol. 8:103-110.
|
|
|
|
|
Daims H (2001). Population Structure and Functional Analyses, by In Situ Techniques, of Nitrifying Bacteria in Wastewater Treatment Plants (Doctoral dissertation, Dissertation Technische Universität München).
|
|
|
|
|
Duncan EG, O'Sullivan CA, Simonsen AK, Roper MM, Peoples MB, Treble K, Whisson K (2017). The nitrification inhibitor 3,4,-dimethylpyrazole phosphate strongly inhibits nitrification in coarse-grained soils containing a low abundance of nitrifying microbiota. Soil Res. 55(1):28-37.
Crossref
|
|
|
|
|
Evdokimova MA, Novoselov SI, Novoselova ES, Mantsurovna A, Kuzminykh AN, Pashkova GI, Marina-Chermnykh OG (2016). The influence of physical properties of sod-podzol soil on its ammonifying and nitrifying capacity. Biol. Med. 8(7):351.
Crossref
|
|
|
|
|
Huson DH, Auch AF, Qi J, Schuster SC (2007). MEGAN analysis of metagenomic data. Genome Res. 17(3):377-386.
Crossref
|
|
|
|
|
Jones JG (1977). The effect of environmental factors on estimated viable and total populations of planktonic bacteria in lakes and experimental inclosures. Freshw. Biol. 7:67-91.
Crossref
|
|
|
|
|
Kämpfer P, Erhart R, Beimfohr C, Böhringer J, Wagner M, Amann R (1996). Characterization of bacterial communities from activated sludge: culture-dependent numerical identification versus in situ identification using group- and genus-specific rRNA-targeted oligonucleotide probes. Microb. Ecol. 32:101-121.
Crossref
|
|
|
|
|
Kirstein K, Bock E (1993). Close genetic relationship between Nitrobacter hamburgensis nitrite oxidoreductase and Escherichia coli nitrate reductases. Archiv. Microbiol. 160(6):447-453.
Crossref
|
|
|
|
|
Kogure K, Simidu U, Taga N (1980). Distribution of viable marine bacteria in neritic seawater around Japan. Can. J. Microbiol. 26:318-323.
Crossref
|
|
|
|
|
Marchesi JR (2012). Metagenomics: current innovations and future trends. Future Microbiol. 7(7):813-814.
Crossref
|
|
|
|
|
Marco D (2011). Metagenomics: current innovations and future trends. Horizon Scientific Press.
|
|
|
|
|
Metzker ML (2010). Sequencing technologies-the next generation. Nat. Rev. Genet. 11(1):31-46.
Crossref
|
|
|
|
|
Poly F, Wertz S, Brothier E, Degrange V (2008). First exploration of Nitrobacter diversity in soils by a PCR cloning-sequencing approach targeting functional gene nxrA. FEMS Microbiol. Ecol. 63(1):132-140.
Crossref
|
|
|
|
|
Pommering-Röser A, Rath G, Koops HP (1996). Phylogenetic diversity within the genus Nitrosomonas. Syst. Appl. Microbiol. 19:344-351.
Crossref
|
|
|
|
|
Purkhold U, Pommerening-Röser A, Juretschko S, Schmid M C, Koops HP, Wagner M (2000). Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66(12):5368-5382.
Crossref
|
|
|
|
|
Qian P, Schoenau JJ (2002). Availability of nitrogen in solid manure amendments with different C: N ratios. Can. J. Soil Sci. 82(2):219-225.
Crossref
|
|
|
|
|
Rotthauwe JH, Witzel KP, Liesack W (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63(12):4704-4712.
|
|
|
|
|
Rughöft S, Herrmann M, Lazar CS, Cesarz S, Levick SR, Trumbore SE, Küsel K (2016). Community composition and abundance of bacterial, archaeal and nitrifying populations in savanna soils on contrasting bedrock material in Kruger National Park, South Africa. Front. Microbiol. 7:1638.
|
|
|
|
|
Saijai S, Ando A, Inukai R, Shinohara M, Ogawa J (2016). Analysis of microbial community and nitrogen transition with enriched nitrifying soil microbes for organic hydroponics. Biosci. Biotechnol. Biochem. 80(11):2247-2254.
Crossref
|
|
|
|
|
Starkenburg, SR, Chain PS, Sayavedra-Soto LA, Hauser L, Larimer FW, Hickey WJ (2006). Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl. Environ. Microbiol. 72(3):2050-2063.
Crossref
|
|
|
|
|
Torsvik V, Goksoyr J, Daae FL (1990). High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787.
|
|
|
|
|
Wertz S, Goyer C, Zebarth BJ, Burton DL, Tatti E, Chantigny MH, Filion M (2013). Effects of temperatures near the freezing point on N2O emissions, denitrification and on the abundance and structure of nitrifying and denitrifying soil communities. FEMS Microbiol. Ecol. 83(1):242-254.
Crossref
|
|
|
|
|
Wertz S, Leigh AK, Grayston SJ (2012). Effects of long-term fertilization of forest soils on potential nitrification and on the abundance and community structure of ammonia oxidizers and nitrite oxidizers. FEMS Microbiol. Ecol. 79(1):142-154.
Crossref
|
|
|
|
|
Willuweit T, Söll P, Müller R (2008). U.S. Patent No. 7,384,777. Washington, DC: U.S. Patent and Trademark Office.
|
|
|
|
|
Winslow CE (1917). The characterization and classification of bacterial types. Science 39(994):77-91
Crossref
|
|
|
|
|
Woese CR, Weisburg WG, Paster BJ, Hahn CM, Tanner RS, Krieg NR,Koops HP, Harms H, Stackebrandt E (1984). The phylogeny of the purple bacteria: The beta subdivision. Syst. Appl. Microbiol. 5:327-336.
Crossref
|
|
|
|
|
Zhou X, Fornara D, Wasson EA, Wang D, Ren G, Christie P, Jia Z (2015). Effects of 44 years of chronic nitrogen fertilization on the soil nitrifying community of permanent grassland. Soil Biol. Biochem. 91:76-83.
Crossref
|
|