Full Length Research Paper
ABSTRACT
This study examined the biodegradability of polystyrene (PS) plastics. Soil samples were collected from Oluku Community in Egor Local Government Area, Benin City, Edo State, Nigeria. Heterotrophic bacteria were enumerated and screened for PS degradation potential. Plastics degrading potential of the isolates was determined by Shake Flask method, degradation of PS plastics was determined by analyzing the formulated PS plastic solution for its additive concentration before and after the degradation process using gas chromatograph with mass spectrometry. Identified bacterial isolates were further characterized using the 16S ribosomal RNA gene. The results from all the parameters indicate that there was active utilization of oxygen and other nutrients available in the test system which is an evidence of PS degradation. The pH had values ranging from 6.5 and 7.4. It was observed that the nutrients and the biochemical oxygen demand decreased considerably with time. There was a reduction in the concentration of bisphenol A (BPA) contingents recorded before (37.04 mg/kg) and after (1.19 mg/kg) the degradation process. The bacterial isolates with codes B1 and B3 belonging to Bacillus while B2 belong to Pseudomonas genera were identified. Two isolates had 99% similarity with Bacillus subtilis strain BS3902 and EU047884.1 respectively, while the third isolate had 100% similarity with Pseudomonas aeruginosa strain KAVKOI. This results shows that the strains have the ability and are able to degrade PS plastics.
Key words: Polystyrene plastics, plastic composted soil, biodegradability, heterotrophic bacteria, molecular characterization.
INTRODUCTION
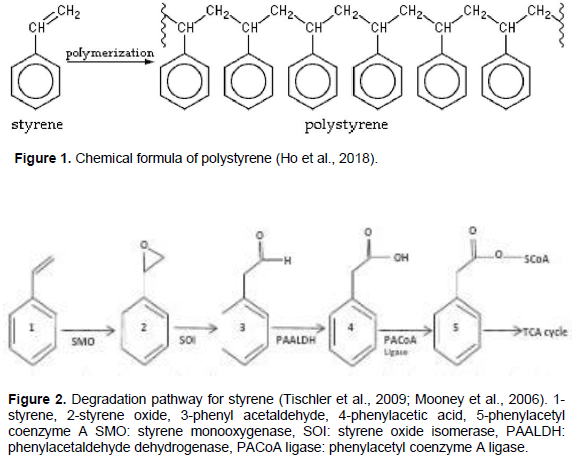
The term “plastics” includes materials composed of various elements such as carbon, hydrogen, oxygen, nitrogen, chlorine, and sulphur. They are produced by the conversion of natural products or by the synthesis from primary chemicals generally coming from crude oil, natural gas, or coal (Coors et al., 2003; Jonsson et al., 2003). The increased use of plastics in day to day consumer applications has resulted in municipal solid waste containing an ever growing fraction of plastic material used for a short time and discarded (Ho et al., 2018). Plastics have taken centre stage in daily life due to its qualities like low weight, durability and low cost as compared to other materials types (Andrady and Neal, 2009). Polystyrene (PS) is a synthetic aromatic polymer with high molecular weight (formula (C8H8)n) made from the monomer styrene (Figure 1) (Ho et al., 2018). Like other plastics, PS is widely used because of its good mechanical properties and relatively low cost (Ho et al., 2018). PS is widely used in construction materials (insulation), packaging foam, food containers, disposable cups, plates, cutleries, cassette boxes, and compact disks (Ho et al., 2018). There is about 21 million tons of PS produced in the world in 2013 (Yang et al., 2015). As a result of such wide use, plastics including PS have accumulated in the environment, causing environmental pollution, human health problems, and ecosystem changes due to their toxicity and recalcitrant compounds. PS materials can be recycled; however, most PS foam ends in landfill (Ho et al., 2018). Plastic pollution affects soil aeration, soil fertility, soil pH, nitrification and the activities of soil fauna and soil flora which act as sentinels in the soil (Atuanya et al., 2016).
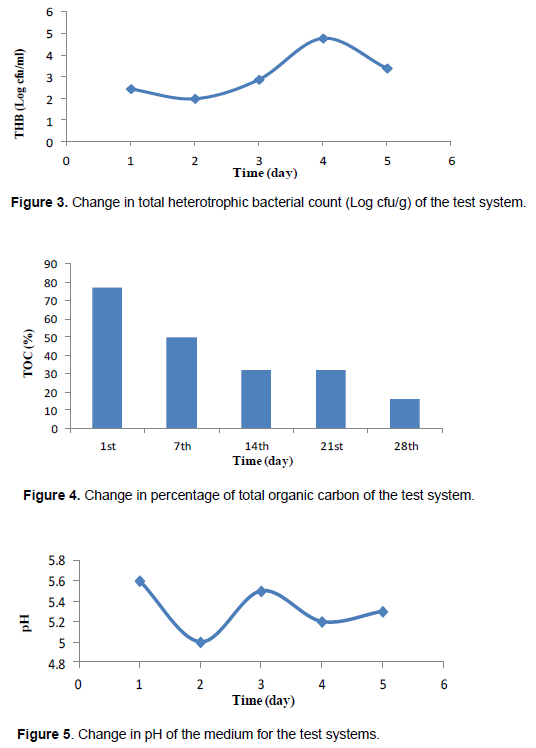
Biodegradation of plastics is the process in which microorganisms (fungi, bacteria, and archaea) degrade them by their extracellular or intracellular enzymes and use the plastics as a substrate for growth (Adamcova and Vaverkova, 2014; Himani et al., 2013; Zheng et al., 2005). PS biodegradation starts when microorganisms begin growing on the surface of PS and secrete their enzymes to degrade the polymer into smaller molecular fragments called oligomer and maybe monomeric units (Zheng et al., 2005). Styrene itself is able to be used as a carbon source for growth by some microorganisms. Rhodococcus ruber has been shown to form biofilms on PS and partially degrade it (Mor and Sivan, 2008). A biofilter consisting of Brevibacillus species has been shown to remove 3 kg of styrene in a day (Motta et al., 2009). The biodegradation rate depends on the thickness and the molecular weight of the plastic (Hwang et al., 2008). In fact, a large number of microorganisms can bring about styrene biodegradation (Baggi et al., 1983). There are several ways of styrene catabolism; however, a predominant pathway involves the oxidation of styrene to phenylacetate, which is then converted via the TCA cycle (Mor and Sivan, 2008). This pathway is as shown in Figure 2.
Biodegradation of PS has been reported in some previous studies. In the literature, few reports describe the microbial utilization of PS as a carbon source (Kaplan et al., 1979; Sielicki et al., 1978). However, there are few reports of microbes degrading PS in the real environment such as landfill, soil, etc. Oikawa et al. (2003) isolated and identified Pseudomonas and Bacillus species for styrene degradation; also Xanthomonas and Sphingobacterium species for PS decomposition by 16 S ribosomal DNA analysis from soil (Sielicki et al., 1978). Four microbial strains have been isolated from garden soil after 8-month buried samples of PS and EPS solution (2%) in chloroform. They were identified as Microbacterium species NA23, Paenibacillus urinalis NA26, Bacillus spp. NB6, and Pseudomonas aeruginosa NB26. They were able to extract some carbon from the complex molecules of PS but the process was very slow and caused no significant chemical changes on the surface (Atiq et al., 2010). Therefore, this study examined biodegradability of polystyrene plastics by bacterial isolates from Plastic Composted waste Soil and Molecular Characterization of Plastic Degrading Bacterial Isolates.
MATERIALS AND METHODS
Sample collection
Soil samples (500 g) were collected from different locations within the waste management landfill site located at Oluku Community, Benin City, Edo State, Nigeria at a depth of 0 to 10 cm with a standard soil auger in plastic bags. The soil samples were homogenized and kept on the laboratory bench to air dry (Atuanya et al., 2012). The soil sample was used for the isolation and enumeration of total heterotrophic bacteria.
Isolation and enumeration of heterotrophic bacteria
Serial dilution of soil sample was made to form 10-4, 10-5 and 10-6 dilutions using normal saline. Total viable heterotrophic bacterial counts were determined. Nutrient agar plates were prepared; the plates were inoculated and were incubated at 37°C for 24 h. Colony counts were taken after incubation and biochemical tests were carried out (Burkhard et al., 2001).
Collection and preparation of polystyrene plastic granules
Waste polystyrene plastics were collected and blended into powder using an industrial grinding machine. The plastic granules was weighed and kept in small white polyethylene bags. This polystyrene granule was used to formulate different polystyrene plastic concentration in a mineral salt medium which was used for biodegradation test (Atuanya et al., 2016).
Screening test for biodegradation potential of polystyrene plastics
Bacterial isolates were screened for the ability to degrade polystyrene plastics using mineral salt medium. 9 ml of the mineral salt medium was dispensed into seven test tubes and sterilized. In each of the test tubes, 0.1 g of plastic at 20 ppm was added to serve as the only source of carbon and energy (Atuanya et al., 2011). Thereafter, all the test tubes were inoculated with two drops of cell suspension of an isolate previously grown in mineral salt medium. The cell suspension was prepared by suspending a loopful of the bacterial isolate from nutrient agar plate into two (2 ml) mineral salt medium. Among the tubes, there was a control which was not inoculated. All the tubes were incubated at room temperature (28±2°C) for 7 days after which the tubes were checked for turbidity which indicated the ability of the isolates to utilize PS plastics as growth source (Ferrara et al., 2006).
Determination of plastics degrading potential of the isolates by shake flask method
A known volume of 150 ml of the mineral salt medium was dispensed into 250 ml conical flask and the test polystyrene (PS) plastic granules were introduced separately into the conical flask after sterilization (Nishida and Tokiwa, 1994). Overnight, broth culture of each isolate was seeded into each flask and incubated on the laboratory bench. The utilization of PS plastics was monitored at two days interval for 10 days by monitoring the bacterial growth measured by viable counts on nutrients agar. The optical density was determined at 620 nm wavelength using Comspec Visible Spectrophotometer, changes in ionic concentration and pH were determined with pH meter (Model Hanna microprocessor P211 pH meter, India) and temperature using temperature meter. Physicochemical analyses were carried out such as pH, total organic carbon, biochemical oxygen demand (BOD), alkalinity analysis, sulphate content, nitrate content and phosphate content to determine the rate of degradability of PS plastic (Brulle et al., 2010).
Determination of plastic degradation
Degradation of the PS plastic granules and the level of degradation was determined using Hewlett Packard HP 5890 series II Gas chromatograph with Mass Spectrometry before and after the degradation process.
Instrumentation and conditions
Hewlett Packard HP 5890 series II Gas chromatograph equipped with an Agilent 7683B injector (Agilent Technologies Santa Clara, CA, USA), A 30 m, 0.25 mm i.d. HP-5MS capillary column (Hewlett – Packard, Palo Alto, CA, USA) coated with 5% phenyl-methylsiloxane (film thickness 0.25 m) and an Agilent 5975 mass selective detector (MSD) was used to separate and quantify the BPA compounds. The samples were injected in the split less mode at an injection temperature of 300°C. The transfer line and ion source temperature was 280 and 200°C. The column temperature was initially held at 40°C for 1 min, raised to 120°C at the rate of 25°C/min, then to 160°C at the rate of 10°C min-1 and finally to 300°C at 5°C min-1, held at final temperature for 15 min. Detector temperature was kept at 280°C. Helium was used as a carries gas at a constant flow rate of ml/min. Mass spectrometry was acquired using the electron ionization (EI) and selective ion monitoring (SIM) mode. A PerkinElmer Gas Chromatograph model Autosystem XL, with Flame Ionization Detector was used for identification of BPA, phthalate, organotin, alkyl phenol and other plastic components by comparison between the retention times of the BPA sample peak and the standard compound. The quantification was done by the internal normalization method. An Elite-5 fused silica capillary column (30 m × 0.25 mm i.d. crossbond 5% diphenyl 95% dimethyl polysiloxane, 0.25 µm film thickness) was used for the GC separation using the following oven temperature program: 150°C (5 min hold) heating to 250°C at 3°C min-1 and heating to 300°C at 10°C min-1 (5 min hold). The injector temperature was 250°C. The injection volume was 1.0 µL (n=3) in the split mode (1:50) (Burkhard et al., 2001).
Molecular characterization of plastic degrading bacterial isolates
DNA extraction
Bacteria in saline were added to 1.5 ml micro centrifuge tube. 450 µl of a 240 mM NaOH, 2.7 mM EDTA, and 74% ethanol solution were added to the tube and mixed gently to give final concentrations of 200 mM NaOH, 2.25 mM EDTA, 61% ethanol. The tube was then heated to 80°C for 10 min and centrifuged at 16,060 ×g for 10 min. The supernatant was removed, and 100 µL of an optimized suspension solution containing 0.1 mM EDTA, 50 Mm Tris-HCI, Ph 8.0, 1% Triton-X-100, and 0.5% Tween-20 was added to solubilize the denatured DNA. DNA was collected by centrifugation at 7200×g for 10 min, washed with 500 µl of 70% ethanol, air dried at room temperature for approximately 3 h and finally dissolved in 50 µl of TE buffer (Brosius et al., 1981).
Polymerase chain reaction procedure
The PCR consist of final volume of 50 µl which included 8 µl DNA and 42 µl reaction cocktail consisting of 5x GoTaq green reaction, 10 Mm of each dNTPs, 10 pmol each 27F: 5´-AGAGTTTGATCMTGGCTCAG-3´ and 1525 R: 5´-AAGGAGGTGWTCCARCC-3´ specific for ? 800 bp conserved domain of the 16S rRNA polymerase. PCR was carried out using the following thermal cycles regime; an initial denaturation at 94°C for 1 min, this was followed by 29 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 1 min and an extension at 72°C for 1.5 min, a final extension at 72°C for 5 min ended the PCR experiment (Brosius et al., 1981).
Agarose gel electrophoresis
Agarose gel was prepared and buffered with 1.5 ml of 0.5x TAE. 10 ml of ethidium bromide was added, mixed and then poured into electrophoretic tank with the comb in place to obtain a gel thickness of about 4 to 5 mm. 10 µl of sample was mixed with 1 µl of the 10x loading dye. DNA samples were loaded and ran. The DNA was viewed using a UV-trans-illuminator (Opere et al., 2013).
Sequencing of the 16S rDNA gene
The purified DNA samples were sequenced at the Bioscience Laboratory, International institute for tropical Agriculture (I.I.T.A), Ibadan, Oyo State with an automated DNA sequencing analyzer (ABI 3730x) using 27F and 1492R primers. Sequence assembly and alignment were carried out using CLC bio software, followed by searching the homology in the Gene Bank using Basic Local Alignment Search Tool (BLAST) program of CLC bio software.
RESULTS AND DISCUSSION
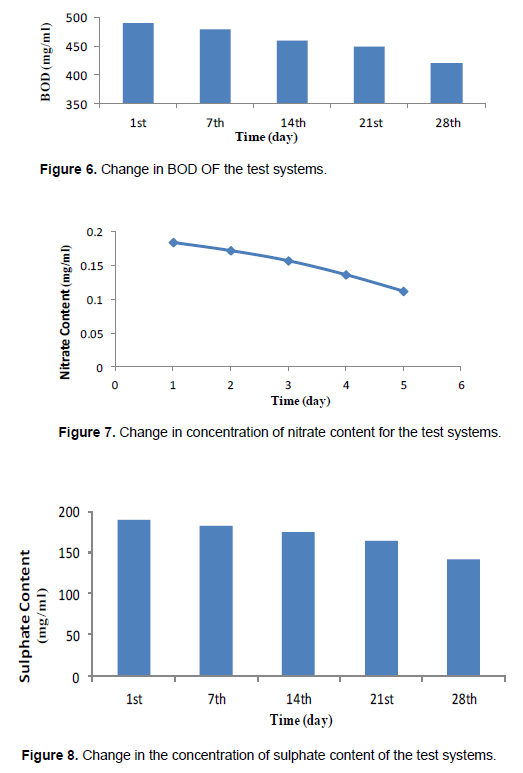
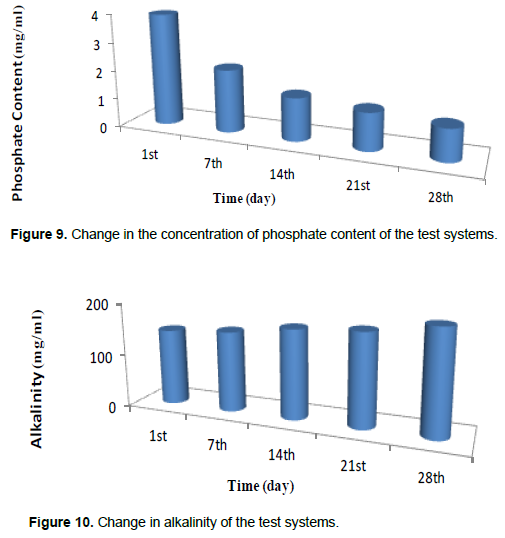
The results from this research showed evidence of polystyrene plastic degradation. All the parameters (Figures 3 to 10) indicate that there was active utilization of oxygen and other nutrients available in the test system. The pH profile obtained generally fell between the optimum range of 6.5 and 7.4 which favors most of the heterotrophic bacterial though the values did not follow a consistent trend as for the other parameters; it was observed that the metabolic products produced by PS plastic utilizing bacteria must have contributed to the fluctuation of the pH readings near neutrality. It was also observed that the nutrients (sulphate, phosphate and nitrate) decreased considerably with time. The decrease is understandable as they are used in the metabolism of microorganism in building biomass. There is correspondence in the utilization of phosphate, sulphate and nitrate indicating their relative importance in cell metabolism as stated by Odum’s combine law. The biochemical oxygen demand (BOD) of the media was also decreasing as the study progressed indicating that the oxygen content in the medium is been utilized by the aerobic bacteria. There was evidence of degradation of polystyrene plastics from the concentration of Bisphenol A (BPA) contingents recorded before (37.04 mg/kg) and after (1.19 mg/kg) the degradation process shown in Table 2. Although there was no complete degradation of the polystyrene plastic, but there was a considerable reduction in the concentration of the BPA contingents, TOC, nitrate, phosphate, and sulphate in the test system (Odokuma and Okpokwasili, 1993).
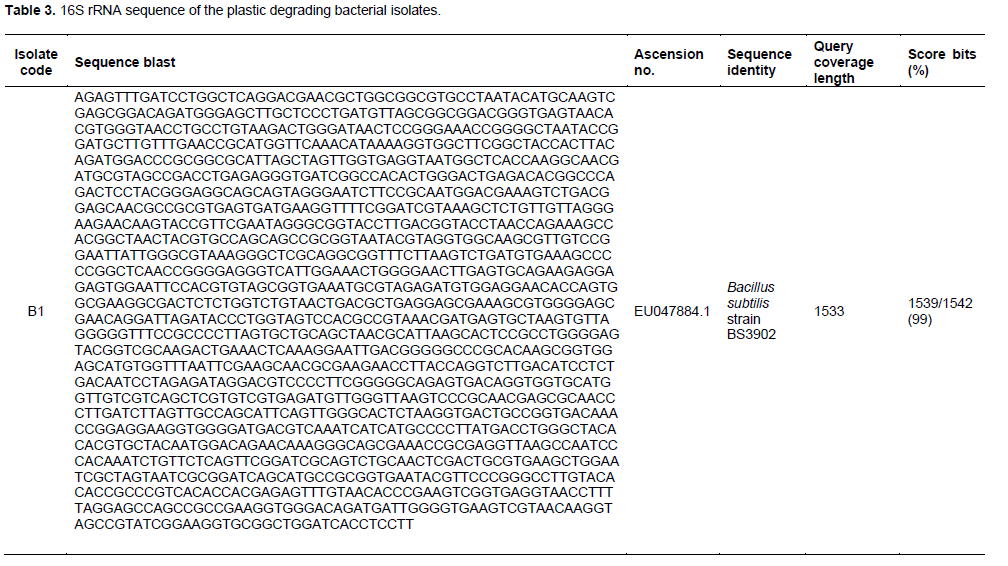
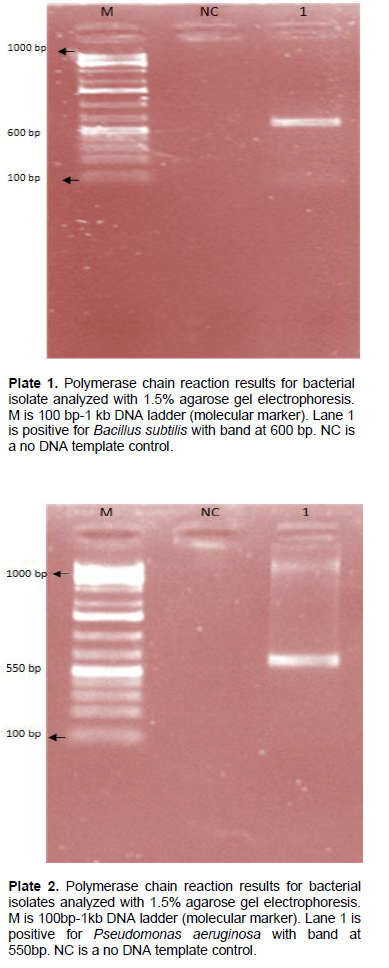
There were three major plastic degrading bacterial isolates of which two were identified as Bacillus spp. and one as Pseudomonas spp. (Table 1) which was further characterized using the 16S ribosomal RNA gene (Table 3). PCR amplification using 16S rRNA gene universal primer set generated amplicons of around 500 bp fragments. This is in line with the results of previous study as theoretically predicted for bacterial family (Opere et al., 2013). Amplicon from the first round of PCR were thereafter used as templates to run a bacterial species level, which generated PCR products of about 600 bp (Plate 1) and 550 bp (Plate 2) in size as predicted for Bacillus and Pseudomonas spp., respectively. BLAST results of the sequences obtained in this study showed an identity query coverage length of 1533, 1532 and2595. It was observed that the isolates from plastic composted soil with codes B1 and B3 belong to Bacillus, while B2 belong to Pseudomonas genera. Two isolates with accession number EU047884.1 and KR967375.1 had 99% similarity with Bacillus subtilis strain BS3902 and B. subtilis strain AER111-2, respectively, while the third isolate had 100% similarity with P. aeruginosa strain KAVKOI with accession number GQ865644.1. It was observed that these strains were able to degrade polystyrene plastics (Opere et al., 2013).
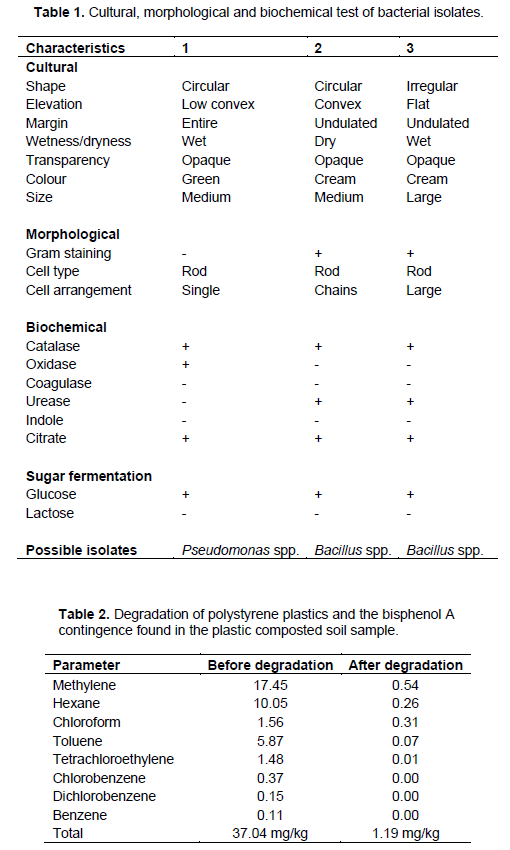


Polystyrene plastics have been found to be susceptible to microbial attack and hence biodegradation or even biodeterioration of these plastics can occur (Okpokwasili and Okorie, 1991). Researchers have reported that P. aeruginosa (Hill, 1978) as the predominant species in petroleum product which is in accordance with this research. This is expected because the genus is commonly found everywhere especially in hydrocarbon polluted area (Fought and Westlake, 1988). The total heterotrophic bacteria differ from those of the hydrocarbon utilizing bacteria when compared. This is due to the ability of the heterotrophic bacteria to withstand stress with time and have resided in the water phase where little nutrient is available. Though there were appropriate bacterial population in the samples, plastic degradation is near impossible if necessary nutrients were not available.
CONCLUSION
The results of the research have shown evidence of polystyrene plastic degradation which is in accordance with previous researches. Time series degradation processes by indigenous microorganisms from the soil have shown to be relatively efficient in the breaking down of plastics products as evidently indicated by the physicochemical analysis.
CONFLICT OF INTERESTS
The authors have not declared any conflict of interests.
REFERENCES
|
Adamcova D, Vaverkova M (2014). Degradation of biodegradable/degradable plastics in municipal solid-waste landfill. Polyer Journal of Environmental Study 23:1071-1078. |
|
|
Andrady AL, Neal MA (2009). Applications and societal benefits of plastics. Philosophical Transaction of the Royal Society of Biologist 364:1977-1984. |
|
|
Atiq N, Safia A, Ali MI (2010). Isolation and identification of polystyrene biodegrading bacteria from soil. African Journal of Microbiological Resource 4:1537-1541. |
|
|
Atuanya EI, Aborisade WT, Nwogu NA (2012). Impact of Plastic Enriched Composting on Soil Structure, Fertility and Growth of Maize Plants. European Journal of Applied Sciences 4(3):105-109. |
|
|
Atuanya EI, Nwogu NA, Akpaje EO (2011). Biodegradation of Polyethylene Film by White-Rot Fungus Pleurotus tuberrgium. Nigerian Journal of Applied Science 29:19-25. |
|
|
Atuanya EI, Udochukwu U, Daveomoregie AO, Inetianbor J (2016). Toxicological effects of plastic composted soil on nitrifying bacteria. British Microbiology Research Journal 13(4):1-7. |
|
|
Ho BT, Timothy KR, Steven L (2018). An overview on biodegradation of polystyrene and modified polystyrene: the microbial approach. Critical Reviews in Biotechnology 38(2):308-320. |
|
|
Baggi G, Boga MM, Catelani D (1983). Styrene catabolism by a strain of Pseudomonas fluorescens. Systematic and Applied Microbiology 4:141-147. |
|
|
Brosius J, Dull TL, Sleeter DD, Noller HF (1981). Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. Journal of Molecular Biology 148:107-127. |
|
|
Brulle F, Morgan AJ, Cocquerelle C, Vandelbulcke F (2010). Transcriptomic underpinning of toxicant-mediated physiological function alterations in three terrestrial invertebrate taxa: A review. Environmental Pollution 158(9):2793-2808. |
|
|
Burkhard S, Bernd MR, Enrico M (2001). The Arabidopsis thaliana ABC transporter AtMRP5 controls root development and stomata movement. Environmental Microbiology Journal 20:1875-1887. |
|
|
Coors A, Jones PD, Giesy JP, Ratte HT (2003). Removal of estrogenic activity from municipal waste landfill leachate assessed with a bioassay based on reporter gene expression. Environmental Science and Technology 37:3430-3434. |
|
|
Ferrara G, Loffredo E, Senesi N (2006). Effects of bisphenol A on the microtubule arrays in root meristematic cells of Pisum sativum. Plant Letters 223:910-916. |
|
|
Fought JM, Westlake DWS (1988). Degradation of Polycyclic Aromatic Hydrocarbon and Aromatic Heterocycles By Pseudomonas species. Canadian Journal of Microbiology 34:1135-1141. |
|
|
Hill EC (1978). Microbial Degradation of Marine Lubricants, Its Detection and Control. Transactions of the Institute of Marine Engineers 90:197-216. |
|
|
Himani B, Richa G, Archana T (2013). Communities of microbial enzymes associated with biodegradation of plastics. Journal of Polymers and the Environment 21:575-579. |
|
|
Hwang JW, Choi CY, Park S (2008). Biodegradation of gaseous styrene by Brevibacillus sp. using a novel agitating biotrickling filter. Biotechnology Letters 30:1207-1212. |
|
|
Jonsson S, Ejlertsson J, Ledin A, Mersiowsky I, Svensson BH (2003). Mono- and diesters from o-phthalic acid in leachates from different European landfills. Water Recourses 37:609-617. |
|
|
Kaplan DL, Roy H, Jim S (1979). Biodegradation of Polystyrene, poly(metnyl methacrylate), and phenol formaldehyde. Applied Environmental Microbiology 38:551-553. |
|
|
Mooney A, Ward PG, O'Connor KE (2006). Microbial degradation of styrene: biochemistry, molecular genetics, and perspective for biotechnical applications. Applied Microbiology and Biotechnology 72:1-10. |
|
|
Mor R, Sivan A (2008). Biofilm formation and partial biodegradation of polystyrene by the actinomycete Rhodococcus ruber: Biodegradation of polystyrene. Biodegradation 19:851-858. |
|
|
Motta O, Proto A, De Carlo F (2009). Utilization of chemically oxidized polystyrene as co-substrate by filamentous fungi. International Journal of Hygiene and Environmental Health 212:61-66. |
|
|
Nishida H, Tokiwa Y (1994) Confirmation of poly(1,3-dioxolan-2-one)-degrading microorganisms in the environment. Chemical Letters 3:421-422. |
|
|
Odokuma LO, Okpokwasili GC (1993). Role of Composition in the Degradability of Oil Spill Dispersants. Waste Management 12:39-43. |
|
|
Oikawa E, Linn KT, Endo T (2003). Isolation and characterization of polystyrene degrading microorganisms for zero emission treatment of expanded polystyrene. Environmental Engineering Research 40:373-379. |
|
|
Okpokwasili GC, Okorie BB (1991). Influence of Physicochemical Stress of Biodegradability of Car Engine Lubricating Oil. International Bioterrorism 27:255-264. |
|
|
Opere BO, Ojo JO, Omonigbehin E, Bamidele M (2013). Antibiotic susceptibility and plasmid profile analysis of pathogenic bacteria isolated from environmental surfaces in public toilets. Transitional Journal of Science and Technology 3(2):22-30. |
|
|
Sielicki M, Focht DD, Martin JP (1978). Microbial degradation of [C14C] polystyrene and 1,3-diphenylbutane. Canadian Journal of Microbiology 24:798-803. |
|
|
Tischler D, Eulberg D, Lakner S (2009). Identification of a novel self-sufficient styrene monooxygenase from Rhodococcus opacus 1CP. Journal of Bacteriology 191:4996-5009. |
|
|
Yang Y, Yang J, Wu WM (2015). Biodegradation and mineralization of polystyrene by plastic-eating mealworms (Part 2): Role of gut microorganisms. Environmental Science and Technology 49:12087-12093. |
|
|
Zheng Y, Yanful EK, Bassi AS (2005). A review of plastic waste biodegradation. Critical Reviews in Biotechnology 25:243-250. |
|
Copyright © 2024 Author(s) retain the copyright of this article.
This article is published under the terms of the Creative Commons Attribution License 4.0